For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPEEHRFVSRRTLVWRRFLRNRPAVVSLVILAALAISCWALPPLLPYSHTDLDYYALQQPPTTTHWFGTN ALGQDILAQTLRGMQKSLLIGVCVAFLSTLIAATAGAIAGYFGAWRSSALMWLVDLLLVVPSFLLIAIVT PRTRESGTLIWLILLLAAFSWMISARIVRGLTMSLREREFVTAARYMGVRNWRIIVRHILPNVASILIID TALNVGLAVLAETSLSFLGFGVQPPDVSLGTLIADGTRSVTTFPWVFLFPAGVLVLIAFCANLVGDGLRD ALDPGVRPPRRRMRRARA*
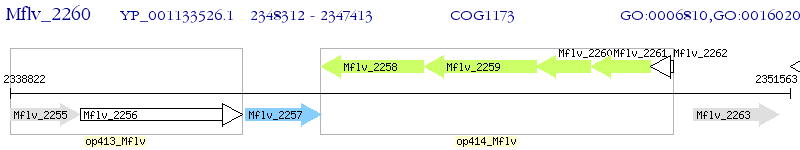
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2260 | - | - | 100% (299) | binding-protein-dependent transport systems inner membrane component |
| M. gilvum PYR-GCK | Mflv_2759 | - | 4e-50 | 37.20% (293) | binding-protein-dependent transport systems inner membrane |
| M. gilvum PYR-GCK | Mflv_0117 | - | 3e-27 | 28.79% (257) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1313c | oppC | 1e-121 | 74.31% (288) | oligopeptide-transport integral membrane protein ABC |
| M. tuberculosis H37Rv | Rv1282c | oppC | 1e-121 | 74.31% (288) | oligopeptide-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01123 | - | 1e-114 | 68.51% (289) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 7e-38 | 33.21% (277) | peptide ABC transporter DppC |
| M. marinum M | MMAR_4137 | oppC | 1e-120 | 74.30% (284) | oligopeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_1432 | oppC | 1e-119 | 73.94% (284) | ABC transporter, permease protein OppC |
| M. smegmatis MC2 155 | MSMEG_4996 | oppC | 1e-130 | 77.51% (289) | ABC transporter, permease protein OppC |
| M. thermoresistible (build 8) | TH_3534 | - | 1e-137 | 82.35% (289) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4000 | oppC | 1e-120 | 74.30% (284) | oligopeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_4431 | - | 1e-150 | 90.41% (292) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2260|M.gilvum_PYR-GCK MSPEE-------------HRFVSRRTLV--WRRFLRNRPAVVSLVILAAL
Mvan_4431|M.vanbaalenii_PYR-1 MTVSAP------------QEFVSRRTLV--WRRFLRNRAAVLSLFVLAAL
TH_3534|M.thermoresistible__bu MTDTASA----ARSGLHTERFVSRRTLV--LRRFLRNRPAVISLAVLALM
MSMEG_4996|M.smegmatis_MC2_155 MTDTSAES---ARSGLHTEEFATRRTLV--FRRFVRNKPAVVSLAVLVLM
Mb1313c|M.bovis_AF2122/97 -----------------MTEFASRRTLV--VRRFLRNRAAVASLAALLLL
Rv1282c|M.tuberculosis_H37Rv -----------------MTEFASRRTLV--VRRFLRNRAAVASLAALLLL
MMAR_4137|M.marinum_M MTDQA-----------TQVEFTSRRTLV--LRRFLRNRAAVASLAVLVVL
MUL_4000|M.ulcerans_Agy99 MTDQA-----------TQVEFTSRRTLV--LRRFLRNRAAVASLAVLVVL
MAV_1432|M.avium_104 MTAQANVDGAHGFSGHEVAGFASRRTLV--LRRFGRNRLAVTSLTLLVLL
MLBr_01123|M.leprae_Br4923 MTLTEG------LSISEKTNFASRKTLV--LRRFAHNRTASVSLMLLVLL
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRSGFFITGSILLTI
:. : * :: :. : * :
Mflv_2260|M.gilvum_PYR-GCK AISCWALPPLLPYS--HTDLDYYALQQPPTTTHWFGTNALGQDILAQTLR
Mvan_4431|M.vanbaalenii_PYR-1 FIGCYALPPLLPYS--HTDLDYYALQQPPSTTHWFGTNALGQDLLAQTLR
TH_3534|M.thermoresistible__bu FVGCYALPPLLPYS--YTDLDYTALQQPPSAEHWFGTNGLGQDILAQTLR
MSMEG_4996|M.smegmatis_MC2_155 FVGCYALPPLLPYS--YTDLDYYALQQPPSPEHWFGTTALGQDILAQVLR
Mb1313c|M.bovis_AF2122/97 FVSAYALPPLLPYS--YDDLDFNALLQPPGTKHWLGTNALGQDLLAQTLR
Rv1282c|M.tuberculosis_H37Rv FVSAYALPPLLPYS--YDDLDFNALLQPPGTKHWLGTNALGQDLLAQTLR
MMAR_4137|M.marinum_M FVGCYALPSVLPYS--YEDLDFDALLQPPSTRHWLGTNALGQDLLAQTLR
MUL_4000|M.ulcerans_Agy99 FVGCYALPSVLPYS--YEDLDFDALLQPPSTRHWLGTNALGQDLLAQTLR
MAV_1432|M.avium_104 FVGCYTLPAVLPYS--YQDLDFDALLQPPNARHWLGTNALGQDLLAQILR
MLBr_01123|M.leprae_Br4923 FVACYLLPSLLPYS--YDDLDFNALLQPPNTRHWLGTNALGQDLLAQTLR
MAB_0428|M.abscessus_ATCC_1997 LVLMALAPQLFTFGKDPRACDLYQARKGISAAHWLGTDLQGCDYYANVVY
: * ::.:. * : . **:** * * *: :
Mflv_2260|M.gilvum_PYR-GCK GMQKSLLIGVCVAFLSTLIAATAGAIAGYFGAWRSSALMWLVDLLLVVPS
Mvan_4431|M.vanbaalenii_PYR-1 GMQKSMLIGVCVAFLSTAIAATVGAVAGYFGAWRSAALMWIVDLLLVVPS
TH_3534|M.thermoresistible__bu GMQKSLLIGVCVAFISTLFAATVGAVAGYFAGWRDRALMWLVDLLLVVPS
MSMEG_4996|M.smegmatis_MC2_155 GMQKSMLIGVCVAFISTIIAATVGSIAGYFGGWRDRTLMWIVDVLLVVPS
Mb1313c|M.bovis_AF2122/97 GMQKSMLIGVCVAVISTGIAATVGAISGYFGGWRDRTLMWVVDLLLVVPS
Rv1282c|M.tuberculosis_H37Rv GMQKSMLIGVCVAVISTGIAATVGAISGYFGGWRDRTLMWVVDLLLVVPS
MMAR_4137|M.marinum_M GMQKSMLIGVFVAVISTGIAATVGAVSGYFGGWRDRALMWLVDLLLVVPS
MUL_4000|M.ulcerans_Agy99 GMQKSMLIGVFVAVISTGIAATVGAVSGYFGGWRDRALMWLVDLLLVVPS
MAV_1432|M.avium_104 GMQKSMLIGVCVAVISTGIAATVGSIAGYFGGWRDRVLMWLVDLLLVVPS
MLBr_01123|M.leprae_Br4923 GMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWVVDLLLVVPS
MAB_0428|M.abscessus_ATCC_1997 GARVSLTIGLIAMVGVLIIGVIVGALAGYFGGIADSALSRLTDVFYGIP-
* : *: **: . . :.. .*:::***. . .* :.*:: :*
Mflv_2260|M.gilvum_PYR-GCK FLLIAIVTPRTRESGTLIWLILLLAAFSWMISARIVRGLTMSLREREFVT
Mvan_4431|M.vanbaalenii_PYR-1 FLLIAIVTPRTRESGTILWLILLLSAFSWMISARIVRGMTMSLREREFVT
TH_3534|M.thermoresistible__bu FILIAIITPRTRDTGTILWLILLLAAFSWMISSRMVRGLTMSLREREFVV
MSMEG_4996|M.smegmatis_MC2_155 FILIAIVTPRLNDADRVFWLIVLLSLFSWMISSRIVRGMTMSLRSREFVV
Mb1313c|M.bovis_AF2122/97 FILIAIVTPRTKNSANIMFLVLLLAGFGWMISSRMVRGMTMSLREREFIR
Rv1282c|M.tuberculosis_H37Rv FILIAIVTPRTKNSANIMFLVLLLAGFGWMISSRMVRGMTMSLREREFIR
MMAR_4137|M.marinum_M FILIAIVTPRTKNSTNMMFLVLLLAGFGWMISSRMVRGMTMSLREREFIR
MUL_4000|M.ulcerans_Agy99 FILIAIVTPRTKNSTNMMFLVLLLAGFGWMISSRMVRGMTMSLREREFIR
MAV_1432|M.avium_104 FILIAIVTPRTKNSANILMLVLLLAGFGWMVSSRMVRGMTMSLREREFIR
MLBr_01123|M.leprae_Br4923 FVLIAIMSPQTKSSDNIMLLVLLLAGFGWMVSSRMVRGMTMSLRKSEFIR
MAB_0428|M.abscessus_ATCC_1997 TILGGMILLSVVGHRTVWSVAGALIAFGWMTSMRLVRSSVMSIKQSDYVQ
:* .:: : : * *.** * *:**. .**::. :::
Mflv_2260|M.gilvum_PYR-GCK AARYMGVRNWRIIVRHILPNVASILIIDTALNVGLAVLAETSLSFLGFGV
Mvan_4431|M.vanbaalenii_PYR-1 AARYMGVSNRRIIVRHILPNVASILIIDTALNVGLAVLAETSLSFLGFGV
TH_3534|M.thermoresistible__bu AARYMGVPHWRIIVRHIIPNVASILIIDTALNVGLAVLAETGLSYLGFGV
MSMEG_4996|M.smegmatis_MC2_155 AARYMGVRSGRIIVRHILPNVASLLIIDTALNVGTAILAETGLSFLGFGI
Mb1313c|M.bovis_AF2122/97 AARYMGVSSRRIIVGHVVPNVASILIIDAALNVAAAILAETGLSFLGFGI
Rv1282c|M.tuberculosis_H37Rv AARYMGVSSRRIIVGHVVPNVASILIIDAALNVAAAILAETGLSFLGFGI
MMAR_4137|M.marinum_M AARYMGVSSRRIITGHVIPNVASILIIDAALNVAAAILAETGLSFLGFGI
MUL_4000|M.ulcerans_Agy99 AARYMGVSSRRIITGHVIPNVASILIIDAALNVAAAILAETGLSFLGFGI
MAV_1432|M.avium_104 AARYMGVSSRRIIVGHVVPNVASILIIDAALNVASAILAETGLSFLGFGV
MLBr_01123|M.leprae_Br4923 AGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSAILAETGLSFLGFGI
MAB_0428|M.abscessus_ATCC_1997 AAIALGAPTWRILLRHILPNALAPVLVYATIAVGSFIAAEATLTYMGIGL
*. :*. *:: *::**. : ::: ::: *. : **: *:::*:*:
Mflv_2260|M.gilvum_PYR-GCK QPPDVSLGTLIADGTRSVTTFPWVFLFPAGVLVLIAFCANLVGDGLRDAL
Mvan_4431|M.vanbaalenii_PYR-1 QPPDVSLGTLIADGTRSVTTFPWVFLFPAGVLVLIVFCANLVGDGLRDAL
TH_3534|M.thermoresistible__bu QRPDVSLGTLIADGTPSVTTFPWVFLFPAGVLVVIILCANLIGDGLRDAF
MSMEG_4996|M.smegmatis_MC2_155 QPPDVSLGTLIADGTGSATTFPWVFLFPAGVLVLIILCSNLVGDGLRDAL
Mb1313c|M.bovis_AF2122/97 QPPDVSLGTLIADGTASATAFPWVFLFPASILVLILVCANLTGDGLRDAL
Rv1282c|M.tuberculosis_H37Rv QPPDVSLGTLIADGTASATAFPWVFLFPASILVLILVCANLTGDGLRDAL
MMAR_4137|M.marinum_M QPPDVSLGTLIADGTQSATTFPWVFLFPAGVLVLILVCANLTGDGLRDAL
MUL_4000|M.ulcerans_Agy99 QPPDVSLGTLIADGTQSATTFPWVFLFPAGVLVLILVCANLTGDGLRDAL
MAV_1432|M.avium_104 QPPDVSLGTLIANGTQSATTFPWVFLFPAGVLVLILVCANLTGDGLRDAL
MLBr_01123|M.leprae_Br4923 KPPDVSLGTLIANGTQSATTFPWVFLFPAGVLVVILVCANLTGDGLRDAF
MAB_0428|M.abscessus_ATCC_1997 ELPEISWGLQINDGQSRFASTPRLVIVPALFLSAAVLGFIMVGDALRDAL
: *::* * * :* :: * :.:.** .* . : **.****:
Mflv_2260|M.gilvum_PYR-GCK DPGVRPPRRRMRRARA-
Mvan_4431|M.vanbaalenii_PYR-1 DPGVRPARRKRDSHR--
TH_3534|M.thermoresistible__bu DPGAGALRRASR-----
MSMEG_4996|M.smegmatis_MC2_155 DPGSGGLRRKSRRKGRR
Mb1313c|M.bovis_AF2122/97 DPASRSLRRGVR-----
Rv1282c|M.tuberculosis_H37Rv DPASRSLRRGVR-----
MMAR_4137|M.marinum_M DPASKTLRSGDR-----
MUL_4000|M.ulcerans_Agy99 DPASKTLRSGDR-----
MAV_1432|M.avium_104 DPGSGPARGGRR-----
MLBr_01123|M.leprae_Br4923 DPVSKHLRR--------
MAB_0428|M.abscessus_ATCC_1997 DPRRR------------
**