For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RRWFVAVALVLFGAFATGCEGVLPDNFALPSQFAGGEPRTLDAATAAAQEFVDRRAAGDYAGVWLMYSRQ VRDRISQSDYAKLSQTCGSSLEKVPVTATGVRLDGETRALVRVKPMGFSVQLPVVYENGEWFMAPSDDFA ADLGKPVDQIIAERRADGRCGDEDLSARLAPPSTSMMPPPPPAEPIPSTGESPSGQCSVDVPGCEGYDEC HPVVTGKRDCFPEDESCSKPGNNFPWCRPLASGDQGTPVVLDCSGQPQVKPERIVLTCADNTIAISKIAW VSWSPDGGATGRGTEFRVECVPNCAQGTAHYSPVTITLTGVAPPDFRYTSATIIDEETGTAESWPMR*
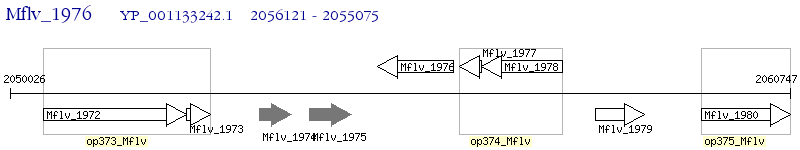
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1976 | - | - | 100% (348) | hypothetical protein Mflv_1976 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0403 | - | 7e-07 | 37.76% (98) | hypothetical protein MAB_0403 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2128 | - | 2e-06 | 36.23% (69) | hypothetical protein MAV_2128 |
| M. smegmatis MC2 155 | MSMEG_5557 | - | 1e-06 | 38.60% (57) | hypothetical protein MSMEG_5557 |
| M. thermoresistible (build 8) | TH_0913 | - | 4e-07 | 33.78% (74) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5557|M.smegmatis_MC2_155 --------------------------------------------------
TH_0913|M.thermoresistible__bu --------------------------------------------------
MAV_2128|M.avium_104 --------------------------------------------------
Mflv_1976|M.gilvum_PYR-GCK MRRWFVAVALVLFGAFATGCEGVLPDNFALPSQFAGGEPRTLDAATAAAQ
MAB_0403|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5557|M.smegmatis_MC2_155 ---------------------------MTRHFAESTAVTSAVTAAAVAAL
TH_0913|M.thermoresistible__bu ---------------------------MIR-------AGAALIGAAALTI
MAV_2128|M.avium_104 ---------------------------MIG-------IVAAVLIGVLTAV
Mflv_1976|M.gilvum_PYR-GCK EFVDRRAAGDYAGVWLMYSRQVRDRISQSDYAKLSQTCGSSLEKVPVTAT
MAB_0403|M.abscessus_ATCC_1997 ---------------------------MTRFVLFAGITLAACVFAGCGAA
:: :
MSMEG_5557|M.smegmatis_MC2_155 MLSAG------------------------------PASAEPVAP-DSGTI
TH_0913|M.thermoresistible__bu LAAAS------------------------------PAAADPAGPADAAAE
MAV_2128|M.avium_104 GMQLT------------------------------RGDHGATAP-AAGPP
Mflv_1976|M.gilvum_PYR-GCK GVRLDGETRALVRVKPMGFSVQLPVVYENGEWFMAPSDDFAADLGKPVDQ
MAB_0403|M.abscessus_ATCC_1997 PNSTSN-------------------------SIRQTSTPAAAASPGRAEP
. ..
MSMEG_5557|M.smegmatis_MC2_155 PENEVYAIGMCYDPEHPLPQRPEVFDYN--CDKTGVLQEMTWTQWGPDGA
TH_0913|M.thermoresistible__bu PHPVSYLIGRCYDPSQPVEERPERILYN--CEGTGLMREMTWTQWDAEGA
MAV_2128|M.avium_104 PTPESYAIPGCYNPSVLPVERPKKLNVLGCASVAVALQDMSWSSWGPQGA
Mflv_1976|M.gilvum_PYR-GCK IIAERRADGRCGDEDLSARLAPPSTSMMPPPPPAEPIPSTGESPSGQCSV
MAB_0403|M.abscessus_ATCC_1997 SRRVVPPGGVYWNPVTQQPEVKPVEIVLTPADAGDMLVDITWDSWTEQQA
: : . .
MSMEG_5557|M.smegmatis_MC2_155 RGTGVDNAIECQPNCAE--------------------------GRRLLNP
TH_0913|M.thermoresistible__bu RGTGIDESVECKPNCAE--------------------------GPRLVNP
MAV_2128|M.avium_104 DGTGRAVFKLCDPNCAT--------------------------GSQLTEP
Mflv_1976|M.gilvum_PYR-GCK DVPGCEGYDECHPVVTGKRDCFPEDESCSKPGNNFPWCRPLASGDQGTPV
MAB_0403|M.abscessus_ATCC_1997 GGHAIRATKDCVPDCAT--------------------------GGTTRRD
. * * : *
MSMEG_5557|M.smegmatis_MC2_155 VVVHAWNPQPSE-SPTCPANARFYSDLTIAYPHDVPPWITPGAEWAPGTD
TH_0913|M.thermoresistible__bu IVVHAWNPRPAD-DDRCPADVQFYADITIAYPQGVPPWIVPGTTWDEGTD
MAV_2128|M.avium_104 VVVHAWNPQPPRREAICQAGLKIFADMILAFPKDVPPPDVQKMN------
Mflv_1976|M.gilvum_PYR-GCK VLDCSGQPQVKPERIVLTCADNTIAISKIAWVSWSPDGGATGRGTEFRVE
MAB_0403|M.abscessus_ATCC_1997 VTLTFHDARSLPDDSGWWQYTKLVITGRDGTSETIDMPVTAR--------
: :.: . . .
MSMEG_5557|M.smegmatis_MC2_155 FTTVDGMPAVHFS----GLTSRCEDQLR--------------------
TH_0913|M.thermoresistible__bu FVIIDGMPAVHFS----DQKPFSCMPLGE-------------------
MAV_2128|M.avium_104 -TQYHGMPAVHFA----NYSSGDTRGTQFIGYTFCN------------
Mflv_1976|M.gilvum_PYR-GCK CVPNCAQGTAHYSPVTITLTGVAPPDFRYTSATIIDEETGTAESWPMR
MAB_0403|M.abscessus_ATCC_1997 ------------------------------------------------