For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVLALLRNRAGATWLLLVAATVLSWAVGADHGTGSLVAVLVLAIAAVKVRLVGLDFMELRHAPIPLRAA FEAYCAGIWVLLSALYLWL
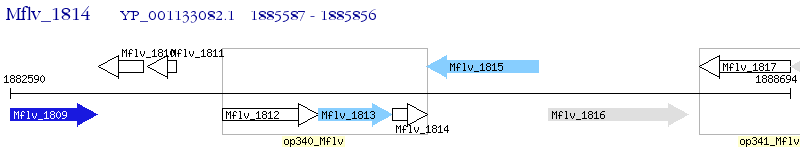
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1814 | - | - | 100% (89) | hypothetical protein Mflv_1814 |
| M. gilvum PYR-GCK | Mflv_4576 | - | 1e-10 | 38.64% (88) | hypothetical protein Mflv_4576 |
| M. gilvum PYR-GCK | Mflv_1249 | - | 4e-07 | 38.46% (78) | hypothetical protein Mflv_1249 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2770 | - | 2e-05 | 31.88% (69) | hypothetical protein MAB_2770 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1935 | - | 2e-25 | 68.49% (73) | hypothetical protein MAV_1935 |
| M. smegmatis MC2 155 | MSMEG_6531 | - | 4e-06 | 34.18% (79) | hypothetical protein MSMEG_6531 |
| M. thermoresistible (build 8) | TH_1787 | - | 4e-08 | 35.44% (79) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4931 | - | 7e-42 | 88.76% (89) | hypothetical protein Mvan_4931 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1814|M.gilvum_PYR-GCK --MTVLALLRNRAG------ATWLLLVAATVLSWAVGADHGTGSL-----
Mvan_4931|M.vanbaalenii_PYR-1 --MTVLSLLKNRAG------ASWLFLVAATVLSWAVGAEHGTGSL-----
MAV_1935|M.avium_104 ------------------------MLIVATVVSYALGADNGTGSV-----
MSMEG_6531|M.smegmatis_MC2_155 --MATRPTTSGES--TRSITWVWLALTAITMGSWWLAPAHITGTVQASTT
TH_1787|M.thermoresistible__bu --MTTDTRHHDDTGAARAMTWAWVALTAITIGSWWLAPAHFKTTVDPSVP
MAB_2770|M.abscessus_ATCC_1997 MIISALPMPARRST-----LVTWLVLTGCTVLSWSESSSGRPSIP-----
* *: *: ..
Mflv_1814|M.gilvum_PYR-GCK VAVLVLAIAAVKVRLVGLDFMELRHAPIPLRAAFEAYCAGIWVLLSALYL
Mvan_4931|M.vanbaalenii_PYR-1 VPVLVLAIAAVKVRLVGLDFMELRHAPVPLRAAFEVYCVGIWALLSALYL
MAV_1935|M.avium_104 MVVVVLAIAAIKVRLVGLDFMELRSAPIPLRLAFEGYCLALWAVLSGLYL
MSMEG_6531|M.smegmatis_MC2_155 ITAVVLVLTFVKCRLIIRHFMEVRTAPRWLGHATDAWLVVLLTTVFVIYL
TH_1787|M.thermoresistible__bu VTVLVLALTLIKSRLVIRYFMEVRTAPRWLKVSTDAWLGALFAVVLIIYL
MAB_2770|M.abscessus_ATCC_1997 VILVMLSLTAIKSALIIASYMEISRAPRWLQMLCGTWVVVVFGMLGYILC
: ::* :: :* *: :**: ** * : : : :
Mflv_1814|M.gilvum_PYR-GCK WL-
Mvan_4931|M.vanbaalenii_PYR-1 WL-
MAV_1935|M.avium_104 FV-
MSMEG_6531|M.smegmatis_MC2_155 F--
TH_1787|M.thermoresistible__bu W--
MAB_2770|M.abscessus_ATCC_1997 VAA