For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADKLSVAIVGSGNISTDLLYKLLRSEWLEPRWMIGIDPESEGLARARKLGLETSHEGVDWLLARDEKPD MVFEATSAYVHRDAAPRYAEAGIRAIDLTPAAIGPGVIPPANLREHLDAPNVNMVTCGGQATIPMVHAVS RVVDVPYAEIVASVSSASAGPGTRANIDEFTKTTSAGVEVIGGARRGKAIIILNPADPPMIMRDTIFCAI PEDADHAAITQSVKDVVAEVQTYVPGYRLLNEPQFDEPSVVNGGNHLVTIFVEVEGAGDYLPPYAGNLDI MTAAAAKVGEEIARERVATSTTGAQA
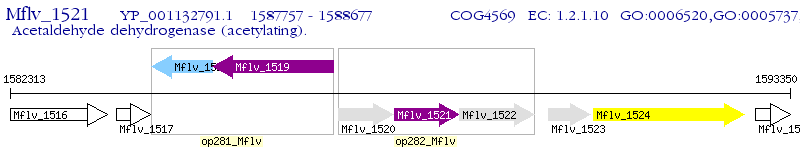
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1521 | - | - | 100% (306) | acetaldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3565c | - | 1e-147 | 85.42% (295) | acetaldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv3535c | - | 1e-147 | 85.42% (295) | acetaldehyde dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0625 | - | 1e-150 | 85.10% (302) | acetaldehyde dehydrogenase |
| M. marinum M | MMAR_5022 | mhpF_1 | 1e-145 | 82.89% (304) | acetaldehyde dehydrogenase, MhpF_1 |
| M. avium 104 | MAV_0626 | - | 1e-148 | 84.33% (300) | acetaldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5939 | - | 1e-158 | 89.90% (307) | acetaldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_0370 | - | 1e-155 | 87.99% (308) | PROBABLE ACETALDEHYDE DEHYDROGENASE (ACETALDEHYDE DEHYDROGENASE |
| M. ulcerans Agy99 | MUL_4096 | mhpF_1 | 1e-145 | 82.89% (304) | acetaldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5235 | - | 1e-161 | 91.50% (306) | acetaldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1521|M.gilvum_PYR-GCK --------------MADKLSVAIVGSGNISTDLLYKLLRSEWLEPRWMIG
Mvan_5235|M.vanbaalenii_PYR-1 --------------MADKKSVAIVGSGNISTDLLYKLLRSEWLEPRWMIG
MSMEG_5939|M.smegmatis_MC2_155 MMSSTKTFEQLGETMPKKSSVAIVGSGNISTDLLYKLLRSEWLEPRWMIG
TH_0370|M.thermoresistible__bu ----------MTATTGRKLQAAIVGSGNISTDLLYKLLRSDWLEPRWMIG
Mb3565c|M.bovis_AF2122/97 --------------MPSKAKVAIVGSGNISTDLLYKLLRSEWLEPRWMVG
Rv3535c|M.tuberculosis_H37Rv --------------MPSKAKVAIVGSGNISTDLLYKLLRSEWLEPRWMVG
MAV_0626|M.avium_104 --------------MPTKAKVAIVGSGNISTDLLYKLLRSDWLEPRWMVG
MMAR_5022|M.marinum_M --------------MPSKASVAIVGSGNISTDLLYKLLRSDWLEPRWMVG
MUL_4096|M.ulcerans_Agy99 --------------MPSKASVAIVGSGNISTDLLYKLLRSDWLEPRWMVG
MAB_0625|M.abscessus_ATCC_1997 --------------MASKASVAIVGSGNISTDLLYKLQRSEWLEPRWMIG
* ..**************** **:*******:*
Mflv_1521|M.gilvum_PYR-GCK IDPESEGLARARKLGLETSHEGVDWLLARDEKPDMVFEATSAYVHRDAAP
Mvan_5235|M.vanbaalenii_PYR-1 IDPESEGLARARKLGLETSHEGVDWLLAQSELPDMVFEATSAYVHKAAAP
MSMEG_5939|M.smegmatis_MC2_155 IDPESEGLARARKLGLETSAEGVDWLLAQSEKPDLVFEATSAYVHRDAAP
TH_0370|M.thermoresistible__bu IDPASEGLARARKLGLETSHEGVDWLFAQDEKPDIVFEATSAYVHREAAP
Mb3565c|M.bovis_AF2122/97 IDPESDGLARAAKLGLETTHEGVDWLLAQPDKPDLVFEATSAYVHRDAAP
Rv3535c|M.tuberculosis_H37Rv IDPESDGLARAAKLGLETTHEGVDWLLAQPDKPDLVFEATSAYVHRDAAP
MAV_0626|M.avium_104 IDPQSEGLARARKLGLETTHEGVDWLLAQPEKPDLVFEATSAYVHRDAAP
MMAR_5022|M.marinum_M IDPQSEGLARARKLGLETTHEGVDWLLAQSDKPDLVFEATSAYVHKAAAP
MUL_4096|M.ulcerans_Agy99 IDPQSEGLARARKLGLETTHEGVDWLVAQSDKPDLVFEATSAYVHKAAAP
MAB_0625|M.abscessus_ATCC_1997 IDPESEGLKRARGFGLETSHEGVDWLLGQDEKPDLVFEATSAYVHKAAAP
*** *:** ** :****: ******..: : **:**********: ***
Mflv_1521|M.gilvum_PYR-GCK RYAEAGIRAIDLTPAAIGPGVIPPANLREHLDAPNVNMVTCGGQATIPMV
Mvan_5235|M.vanbaalenii_PYR-1 RYAEAGIRAIDLTPAAVGPGVIPPANLRAHLDAPNVNMVTCGGQATIPMV
MSMEG_5939|M.smegmatis_MC2_155 RYEEAGIRAIDLTPAAVGPGVVPPANLRDHLDAPNVNMVTCGGQATIPIV
TH_0370|M.thermoresistible__bu RYAEAGIRAIDLTPAAVGPGVIPPANLREHLDAPNVNMVTCGGQATIPIV
Mb3565c|M.bovis_AF2122/97 KYAEAGIRAIDLTPAAVGPAVIPPANLREHLDAPNVNMITCGGQATIPIV
Rv3535c|M.tuberculosis_H37Rv KYAEAGIRAIDLTPAAVGPAVIPPANLREHLDAPNVNMITCGGQATIPIV
MAV_0626|M.avium_104 KYEAAGIRAIDLTPAAVGPAVIPPANLRQHLDAPNVNMITCGGQATIPIV
MMAR_5022|M.marinum_M KYAAAGIRAIDLTPAAVGPAVVPPANLREHLDAPNVNMITCGGQATIPIV
MUL_4096|M.ulcerans_Agy99 KYAAAGIRAIDLTPAAVGPAVVPPANLREHLDAPNVNMITCGGQATIPIV
MAB_0625|M.abscessus_ATCC_1997 RYEEAGIRAIDLTPAAVGPAVVPPANLRAHLDAPNVNMITCGGQATIPIV
:* ************:**.*:****** *********:*********:*
Mflv_1521|M.gilvum_PYR-GCK HAVSRVVD-----VPYAEIVASVSSASAGPGTRANIDEFTKTTSAGVEVI
Mvan_5235|M.vanbaalenii_PYR-1 YAVSRVVE-----VPYAEIVASVSSASAGPGTRANIDEFTKTTSAGVQNI
MSMEG_5939|M.smegmatis_MC2_155 HAVSRVVD-----VPYAEIVASVSSASAGPGTRANIDEFTKTTSAGVQNI
TH_0370|M.thermoresistible__bu HAVVRAVAAHGGTVPYAEIVASVASVSAGPGTRANIDEFTKTTSKGVETI
Mb3565c|M.bovis_AF2122/97 YAVSRIVE-----VPYAEIVASVASVSAGPGTRANIDEFTKTTARGVQTI
Rv3535c|M.tuberculosis_H37Rv YAVSRIVE-----VPYAEIVASVASVSAGPGTRANIDEFTKTTARGVQTI
MAV_0626|M.avium_104 YAVSRVVE-----VPYAEIVASVASVSAGPGTRANIDEFTKTTSRGVETI
MMAR_5022|M.marinum_M YAVSRAVADLGG-VPYAEIVASVASVSAGPGTRANIDEFTKTTSKGVETI
MUL_4096|M.ulcerans_Agy99 YAVSRAVADLGG-VPYAEIVASVASVSAGPGTRANIDEFTKTTSKGVETI
MAB_0625|M.abscessus_ATCC_1997 YAVSRVVE-----VPYAEIVASVASVSAGPGTRANIDEFTKTTSKGVEVI
:** * * **********:*.*****************: **: *
Mflv_1521|M.gilvum_PYR-GCK GGARRGKAIIILNPADPPMIMRDTIFCAIPEDADHAAITQSVKDVVAEVQ
Mvan_5235|M.vanbaalenii_PYR-1 GGAQRGKAIIILNPAEPPMIMRDTIFCAIPEHADHAAITQSIKDVVAEVQ
MSMEG_5939|M.smegmatis_MC2_155 GGAQRGKAIIVLNPAEPPMIMRDTIFCAIPEGADHDAITQSIKDVVAEVQ
TH_0370|M.thermoresistible__bu GGAQRGKAIIILNPAEPPMIMRDTIFCAIPEDADHDAITESIHDVVAEVQ
Mb3565c|M.bovis_AF2122/97 GGAARGKAIIILNPADPPMIMRDTIFCAIPTDADREAIAASIHDVVKEVQ
Rv3535c|M.tuberculosis_H37Rv GGAARGKAIIILNPADPPMIMRDTIFCAIPTDADREAIAASIHDVVKEVQ
MAV_0626|M.avium_104 GGAKRGKAIIILNPADPPMIMRDTIFCAIPEDADRDAIAQSIHDVVKEVQ
MMAR_5022|M.marinum_M GGARRGKAIIILNPADPPMIMRDTIFCAIPEDADRDAIAASIHEVVAQVQ
MUL_4096|M.ulcerans_Agy99 GGARRGKAIIILNPADPPMIMRDTIFCAIPEDADRDAIAASIHEVVAQVQ
MAB_0625|M.abscessus_ATCC_1997 GGAKRGKAIIILNPADPPMIMRDTIFCAIPEDADRAAIAASIHDVVSQVQ
*** ******:****:************** **: **: *:::** :**
Mflv_1521|M.gilvum_PYR-GCK TYVPGYRLLNEPQFDEPSVVNGGNHLVTIFVEVEGAGDYLPPYAGNLDIM
Mvan_5235|M.vanbaalenii_PYR-1 TYVPGYRLLNEPQFDEPSVVNGGNHVVTVFVEVEGAGDYLPPYAGNLDIM
MSMEG_5939|M.smegmatis_MC2_155 TYVPGYRLLNEPQFDEPSVVNGGNHLVTTFVEVEGAGDYLPPYAGNLDIM
TH_0370|M.thermoresistible__bu TYVPGYRLLNEPQFDEPSVVNGGNHLVTTFIEVEGAGDYLPPYAGNLDIM
Mb3565c|M.bovis_AF2122/97 TYVPGYRLLNEPQFDEPSINSGGQALVTTFVEVEGAGDYLPPYAGNLDIM
Rv3535c|M.tuberculosis_H37Rv TYVPGYRLLNEPQFDEPSINSGGQALVTTFVEVEGAGDYLPPYAGNLDIM
MAV_0626|M.avium_104 SYVPGYRLLNEPQFDDPSLNSGGQALVTTFVEVEGAGDYLPPYAGNLDIM
MMAR_5022|M.marinum_M TYVPGYRLLNEPQFDEPSINSGGQAVVTTFVEVEGAGDYLPPYAGNLDIM
MUL_4096|M.ulcerans_Agy99 TYVPGYRLLNEPQFDEPSINSGGQAVVTTFVEVEGAGDYLPPYAGNLDIM
MAB_0625|M.abscessus_ATCC_1997 QYVPGYRLLNEPQFDEPSVVNGGNHVVTTFVEVEGAGDFLPPYAGNLDIM
**************:**: .**: :** *:*******:***********
Mflv_1521|M.gilvum_PYR-GCK TAAAAKVGEEIARERVATS-TTGAQA
Mvan_5235|M.vanbaalenii_PYR-1 TAAATKVGEEIAKESLAAT-AGGAQA
MSMEG_5939|M.smegmatis_MC2_155 TAAATKVGEEIAKKSAEASLASGAQA
TH_0370|M.thermoresistible__bu TAAATKVGEEIARESLTAT--AGGQA
Mb3565c|M.bovis_AF2122/97 TAAATKVGEEIAKETL-VVGGAR---
Rv3535c|M.tuberculosis_H37Rv TAAATKVGEEIAKETL-VVGGAR---
MAV_0626|M.avium_104 TAAATKVGEEIAKETLSVAGGTR---
MMAR_5022|M.marinum_M TAAATKVGEEIAMQTLAVSGGKR---
MUL_4096|M.ulcerans_Agy99 TAAATKVGEEIAKETLAVSGGKR---
MAB_0625|M.abscessus_ATCC_1997 TAAATKVGEEIAKELLSAKVS-----
****:******* : .