For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AKNSKIVKNERRLVLVARHAERRAELKAIISSPSTPADARAAAQSELNRQPRDASPVRVRNRDAVDGRPR GHLRKFGLSRMRVRELAHRGQLPGVRKSSW*
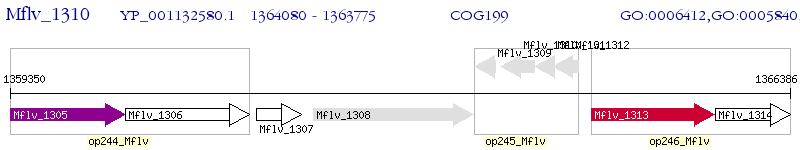
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1310 | rpsN | - | 100% (101) | 30S ribosomal protein S14 |
| M. gilvum PYR-GCK | Mflv_5034 | rpsN | 2e-09 | 46.77% (62) | 30S ribosomal protein S14 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2082c | rpsN | 3e-37 | 71.29% (101) | 30S ribosomal protein S14 |
| M. tuberculosis H37Rv | Rv2056c | rpsN | 3e-37 | 71.29% (101) | 30S ribosomal protein S14 |
| M. leprae Br4923 | MLBr_01846 | rpsN | 3e-10 | 48.39% (62) | 30S ribosomal protein S14 |
| M. abscessus ATCC 19977 | MAB_0332c | rpsN | 4e-39 | 75.25% (101) | 30S ribosomal protein S14 |
| M. marinum M | MMAR_0290 | rpsN | 5e-38 | 73.27% (101) | ribosomal protein S14 RpsN2 |
| M. avium 104 | MAV_4452 | rpsN | 6e-10 | 50.00% (62) | 30S ribosomal protein S14 |
| M. smegmatis MC2 155 | MSMEG_6066 | rpsN | 5e-38 | 73.27% (101) | 30S ribosomal protein S14 |
| M. thermoresistible (build 8) | TH_0310 | - | 3e-09 | 46.77% (62) | PROBABLE 30S RIBOSOMAL PROTEIN S14 RPSN1 |
| M. ulcerans Agy99 | MUL_4833 | rpsN | 4e-38 | 73.27% (101) | ribosomal protein S14 RpsN2 |
| M. vanbaalenii PYR-1 | Mvan_5491 | rpsN | 5e-46 | 87.13% (101) | 30S ribosomal protein S14 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1310|M.gilvum_PYR-GCK MAKNSKIVKNERRLVLVARHAERRAELKAIISSPSTPADARAAAQSELNR
Mvan_5491|M.vanbaalenii_PYR-1 MAKKSKIVQNERRREIVARHAERRAELKAIIKSPSTTPEARVAAQSELNR
Mb2082c|M.bovis_AF2122/97 MAKKSKIVKNQRRAATVARYASRRTALKDIIRSPSSAPEQRSTAQRALAR
Rv2056c|M.tuberculosis_H37Rv MAKKSKIVKNQRRAATVARYASRRTALKDIIRSPSSAPEQRSTAQRALAR
MMAR_0290|M.marinum_M MAKKSKIVKNNKRRDIVARYAQRRAELKQIIQSPTSSYEQRLDAQRALSR
MUL_4833|M.ulcerans_Agy99 MAKKSKIVKNNKRRDIVARYAQRRAELKQIIQSPTSSYEQRLDAQRALSR
MAB_0332c|M.abscessus_ATCC_199 MAKKSKIAKNEQRRAIVERYAERRAELKEIIRSPASSPEQRVAAQSELNR
MSMEG_6066|M.smegmatis_MC2_155 MAKKSKIVKNEQRRELVQRYAERRAELKRTIRDPASSPERRAAAVSALQR
MLBr_01846|M.leprae_Br4923 MAKKALVNKAA--------------------------------------R
MAV_4452|M.avium_104 MAKKALVNKAA--------------------------------------R
TH_0310|M.thermoresistible__bu MAKKALIHKAQ--------------------------------------R
***:: : : *
Mflv_1310|M.gilvum_PYR-GCK QPRDASPVRVRNRDAVDGRPRGHLRKFGLSRMRVRELAHRGQLPGVRKSS
Mvan_5491|M.vanbaalenii_PYR-1 QPRDASPVRVRNRDSVDGRPRGHLRKFGLSRVRVRELAHQGQLPGVRKSS
Mb2082c|M.bovis_AF2122/97 QPRDASPVRLRNRDAIDGRPRGHLRKFGLSRVRVRQLAHDGHLPGVRKAS
Rv2056c|M.tuberculosis_H37Rv QPRDASPVRLRNRDAIDGRPRGHLRKFGLSRVRVRQLAHDGHLPGVRKAS
MMAR_0290|M.marinum_M QPRDASAVRLRNRDAIDGRPRGHLRKFGLSRVRVRELAHAGQLPGVRKAS
MUL_4833|M.ulcerans_Agy99 QPRDASAVRLRNRDAIDGRPRGHLRKFGLSRVRVRELAHAGQLPGVRKAS
MAB_0332c|M.abscessus_ATCC_199 QPRDASAVRLRNRDAVDGRPRGYLRKFGLSRVRVRELVHDGSLPGVRKAS
MSMEG_6066|M.smegmatis_MC2_155 LPRDSSPVRLRNRDVVDGRPRGHLRKFGLSRVRVREMAHRGELPGVRKAS
MLBr_01846|M.leprae_Br4923 KPK--FTVRGYTRCSKCGRPRAVFRKFGLCRICLREMAHAGELPGVQKSS
MAV_4452|M.avium_104 KPK--FAVRGYTRCNKCGRPRAVFRKFGLCRICLREMAHAGELPGVQKSS
TH_0310|M.thermoresistible__bu KPK--FKVRAYTRCNKCGRPRSVYRKFGLCRICLREMAHAGELPGVQKAS
*: ** .* ****. *****.*: :*::.* * ****:*:*
Mflv_1310|M.gilvum_PYR-GCK W
Mvan_5491|M.vanbaalenii_PYR-1 W
Mb2082c|M.bovis_AF2122/97 W
Rv2056c|M.tuberculosis_H37Rv W
MMAR_0290|M.marinum_M W
MUL_4833|M.ulcerans_Agy99 W
MAB_0332c|M.abscessus_ATCC_199 W
MSMEG_6066|M.smegmatis_MC2_155 W
MLBr_01846|M.leprae_Br4923 W
MAV_4452|M.avium_104 W
TH_0310|M.thermoresistible__bu W
*