For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQPGGQPDMSALLAQAQQVQQQLMEAQEALANAEVHGQAGGGLVQVTMKGSGEVVGVSIDPKVVDPSDVE TLQDLIVGAIADAAKQVTILAHDKLGPLAGGMGGLGLPGM
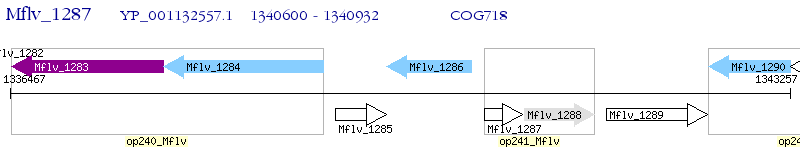
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1287 | - | - | 100% (110) | hypothetical protein Mflv_1287 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3743c | - | 1e-39 | 67.50% (120) | hypothetical protein Mb3743c |
| M. tuberculosis H37Rv | Rv3716c | - | 1e-39 | 67.50% (120) | hypothetical protein Rv3716c |
| M. leprae Br4923 | MLBr_02330 | - | 7e-38 | 73.53% (102) | hypothetical protein MLBr_02330 |
| M. abscessus ATCC 19977 | MAB_0319 | - | 4e-39 | 68.42% (114) | hypothetical protein MAB_0319 |
| M. marinum M | MMAR_5232 | - | 9e-39 | 71.56% (109) | hypothetical protein MMAR_5232 |
| M. avium 104 | MAV_0387 | - | 6e-38 | 78.95% (95) | hypothetical protein MAV_0387 |
| M. smegmatis MC2 155 | MSMEG_6280 | - | 3e-46 | 87.25% (102) | hypothetical protein MSMEG_6280 |
| M. thermoresistible (build 8) | TH_2288 | - | 8e-48 | 91.18% (102) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4307 | - | 7e-39 | 71.56% (109) | hypothetical protein MUL_4307 |
| M. vanbaalenii PYR-1 | Mvan_5528 | - | 7e-55 | 94.55% (110) | hypothetical protein Mvan_5528 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3743c|M.bovis_AF2122/97 MQPGG--DMSALLAQAQQMQQKLLEAQQQLANSEVHGQAGGGLVKVVVKG
Rv3716c|M.tuberculosis_H37Rv MQPGG--DMSALLAQAQQMQQKLLEAQQQLANSEVHGQAGGGLVKVVVKG
MMAR_5232|M.marinum_M MQPGG--DMSALLAQAQQMQQKLLEAQQQLANSEVHGQAGGGLVQVAVRG
MUL_4307|M.ulcerans_Agy99 MQPGG--DMSALLAQAQQMQQKLLEAQQQLANSEVHGQAGGGLVQVAVRG
MLBr_02330|M.leprae_Br4923 MQPGG--DMSALLAQAQQMQQKLLETQQQLANAQVHGQGGGGLVEVVVKG
MAV_0387|M.avium_104 --------MSALLAQAQQMQQKLMEAQQQLANAEVHGQAGGGLVKVVVKG
Mflv_1287|M.gilvum_PYR-GCK MQPGGQPDMSALLAQAQQVQQQLMEAQEALANAEVHGQAGGGLVQVTMKG
Mvan_5528|M.vanbaalenii_PYR-1 MQPGGQPDMSALLAQAQQVQQQLMEAQEALANAEVNGQAGGGLVQVTMRG
TH_2288|M.thermoresistible__bu --------MSALLAQAQQVQQQLMEAQEALANSEVHGQAGGGLVQVTMKG
MSMEG_6280|M.smegmatis_MC2_155 --------MSALLAQAQQVQQQLMEAQEALANAEVHGQAGGGLVQVTVKG
MAB_0319|M.abscessus_ATCC_1997 MQPGGAPDMSALLAQAQQMQQQLMAAQAQIAAAEVTGESGGGLVRITGKG
**********:**:*: :* :* ::* *:.*****.:. :*
Mb3743c|M.bovis_AF2122/97 SGEVIGVTIDPKVVDPDDIETLQDLIVGAMRDASQQVTKMAQERLGALAG
Rv3716c|M.tuberculosis_H37Rv SGEVIGVTIDPKVVDPDDIETLQDLIVGAMRDASQQVTKMAQERLGALAG
MMAR_5232|M.marinum_M SGELVGVTIDPKVVDRDDIETLQDLIVGAMRDASQQVTKMAQERLGALAG
MUL_4307|M.ulcerans_Agy99 SGELVGVTIDPKVVDRDDIETLQDLIVGAMRDASQQVTKMAQERLGALAG
MLBr_02330|M.leprae_Br4923 SGEVVSVAIDPKVVDPGDIETLQDLIVGAMADASKQVTKLAQERLGALTS
MAV_0387|M.avium_104 SGEVVAVKIDPSVVDPSDVETLQDLVVGAMADASKQVTRMAQERLGSLAG
Mflv_1287|M.gilvum_PYR-GCK SGEVVGVSIDPKVVDPSDVETLQDLIVGAIADAAKQVTILAHDKLGPLAG
Mvan_5528|M.vanbaalenii_PYR-1 SGEVVAVRIDPKVVDPSDVETLQDLIVGAVADAAKQVTILAHDRLGPLAG
TH_2288|M.thermoresistible__bu SGEVVAVTIDPKVVDPDDVETLQDLIVGAIADASKQVTILAQDRLGPLAG
MSMEG_6280|M.smegmatis_MC2_155 SGEVVSVQIDPKVVDPDDVETLQDLIVGALSDASKQVTIMAQSRLGPLAG
MAB_0319|M.abscessus_ATCC_1997 SGEVTSVQIDPKIVDPEDVETLQDLIIGALADLTSKTQELASQRLGPLAG
***: .* ***.:** *:******::**: * :.:. :* .:**.*:.
Mb3743c|M.bovis_AF2122/97 AMR-PPAPPAAP--PGAPGMP------GMPGMPGAPGAPPVPGI------
Rv3716c|M.tuberculosis_H37Rv AMR-PPAPPAAP--PGAPGMP------GMPGMPGAPGAPPVPGI------
MMAR_5232|M.marinum_M AMR-PPGAPGAPGAPGAPGMPPRPGAPGMPGMPGAPGAPGMPGMPGAPGV
MUL_4307|M.ulcerans_Agy99 AMR-PPGAPGAPGAPGAPGMPPRPGAPGMPGMPGAPGAPGMPGMPGAPGV
MLBr_02330|M.leprae_Br4923 AMR-PTAPPPTP-----P-----------TYMAGT---------------
MAV_0387|M.avium_104 GFGAPPGQPPAP-------------------APGV---------------
Mflv_1287|M.gilvum_PYR-GCK GMG-GLGLPGM---------------------------------------
Mvan_5528|M.vanbaalenii_PYR-1 GMG-GLGLPGM---------------------------------------
TH_2288|M.thermoresistible__bu GMS-GFGLPGM---------------------------------------
MSMEG_6280|M.smegmatis_MC2_155 GLG-GFGLPGM---------------------------------------
MAB_0319|M.abscessus_ATCC_1997 GLGDLGGGLGLP---------------------GV---------------
.: .