For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRTLGRTRRACYGPPMGNPRRVLLAVLAALAVLLTACGSSGPGNSVKDSGVLRVGTEGVYAPFSFHDPAT GQLTGYDVDVARAVGEKLGVNVEFVETPWDSIFAALEANRFDVVANQVTITPERQQKYDLSEPYSIGEGV IVTRADDDSITSLDDLRGRRAAQSTTSNWAQVARDAGAQIESVEGLTQAMALLSQGRVDVVVNDSLSIYA YLAETGDTAVKIAGTTGEKSEQGFAARKNSGLLPDLNTAVEELKADGTMAEISQKYLKTNASGGQDTPQA QPRSTLQLVLDNLWPLAKAALTMTIPLTIISFVIGLVIALGVALARLSSNVVLSNLARFYISVIRGTPLL VQLFIVFFALPEFGVKIDPFPAAVIAFSLNVGGYAAEIIRSAIQSIPKGQWEAAETIGLNYVGTLRRIIL PQATRVAVPPLSNTLISLVKDTSLASTILVTELLRQAQIIAAPTFEFFALYGTAAVYYWVICLVLSFGQA RFEHRLERYVAR*
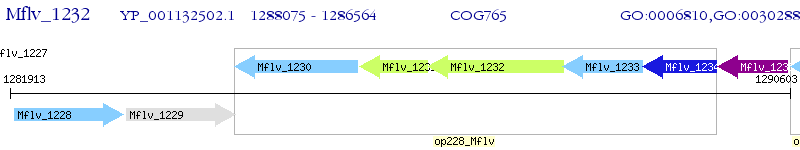
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1232 | - | - | 100% (503) | polar amino acid ABC transporter, inner membrane subunit |
| M. gilvum PYR-GCK | Mflv_3164 | - | 5e-67 | 32.99% (491) | polar amino acid ABC transporter, inner membrane subunit |
| M. gilvum PYR-GCK | Mflv_0858 | - | 2e-37 | 29.80% (443) | polar amino acid ABC transporter, inner membrane subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0419c | glnH | 1e-14 | 29.55% (247) | glutamine-binding lipoprotein |
| M. tuberculosis H37Rv | Rv0411c | glnH | 1e-14 | 29.55% (247) | glutamine-binding lipoprotein |
| M. leprae Br4923 | MLBr_00303 | glnH | 7e-13 | 26.75% (228) | putative glutamine-binding protein |
| M. abscessus ATCC 19977 | MAB_0277c | - | 0.0 | 69.94% (489) | amino acid ABC transporter permease |
| M. marinum M | MMAR_0061 | glnQ | 2e-37 | 27.29% (502) | ABC transporter permease protein GlnQ |
| M. avium 104 | MAV_0059 | - | 2e-34 | 27.48% (473) | amino acid ABC transporter, permease protein, |
| M. smegmatis MC2 155 | MSMEG_6307 | - | 0.0 | 78.32% (475) | glutamine-binding periplasmic protein/glutamine transport |
| M. thermoresistible (build 8) | TH_0515 | - | 5e-37 | 28.37% (497) | amino acid ABC transporter, permease protein, 3-TM region, |
| M. ulcerans Agy99 | MUL_2809 | glnH | 2e-15 | 30.70% (228) | glutamine-binding lipoprotein GlnH |
| M. vanbaalenii PYR-1 | Mvan_5576 | - | 0.0 | 93.64% (503) | polar amino acid ABC transporter, inner membrane subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1232|M.gilvum_PYR-GCK --------------------------MSRTLGRTRRACYGPPMGNPRRVL
Mvan_5576|M.vanbaalenii_PYR-1 --------------------------MSRTLQLTPPGCYGPAMRYPRTVL
MSMEG_6307|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0277c|M.abscessus_ATCC_199 ------------------------------------------------MF
MMAR_0061|M.marinum_M ----------MLAVSATILIVLG--LALAAPALADHNQCAPAGVDSAMVL
MAV_0059|M.avium_104 MGRRGTRRGKLLALTATVLMVCGSSALLAPPAAADRNQCAPAGPKSAAVL
TH_0515|M.thermoresistible__bu --VTRAAVVAVIALLVGSLTAGG--TVLAAPAAATVDQCAPPGVQSASPL
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
Mflv_1232|M.gilvum_PYR-GCK LAVLAALAVLLTACGSSG--PGNSVKDSGV--LRVGTEGVYAPFSFHDP-
Mvan_5576|M.vanbaalenii_PYR-1 LAVLAAIAVLLTACGSSG--SGNPVESSGV--LRVGTEGVYAPFSYHDP-
MSMEG_6307|M.smegmatis_MC2_155 --------MLLAGCGPAANDSDEPIKAAGV--LRVGTEGVYAPFSYHDP-
MAB_0277c|M.abscessus_ATCC_199 RAIVLALLALVAVAGCAA-----PTQEP----LRVGTEGVYAPFSFHDG-
MMAR_0061|M.marinum_M PADLSSATTAGSDEDVQTTAGVVPLSSVNIGDLGLGTPGVLTAGTLSDAP
MAV_0059|M.avium_104 PENLTRGGSMS-QPDEHTTPTVEPLSSVRIDTLGLGTPGVLTVGTLSGAP
TH_0515|M.thermoresistible__bu PTNLAAAEAGP-GVDRYTTDGVVPLDNIDPSALRLRVPGVLTVGTLSDAP
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
Mflv_1232|M.gilvum_PYR-GCK ------ATGQLTGYDVDVARAVGEKLGVNVEFVETPWDSIFAALEANRFD
Mvan_5576|M.vanbaalenii_PYR-1 ------ATGQLTGYDVDVARAVGEKLGVNVEFVETPWDSIFAALEANRFD
MSMEG_6307|M.smegmatis_MC2_155 ------ATNELTGYDIDVVKAVADKIGVQIQFVETPWDSIFAALEANRFD
MAB_0277c|M.abscessus_ATCC_199 ------HTGELTGYDVDVARAVGEKLGRPVEFVEIPWDAIFAGLDAERFD
MMAR_0061|M.marinum_M PNTCINSTGQFTGFDNELLRAIAAKLGLQVRFVGTDFSGLLAQVAARRFD
MAV_0059|M.avium_104 PNVCITPTGQYSGFDNQLLRAIAGKLGLQVRFVGTDFAGLLAQVASRRFD
TH_0515|M.thermoresistible__bu PSICINSLGEFTGFDNELLRAIAAKLGLQISFVGTEFSALLAQVASGRFD
Mb0419c|M.bovis_AF2122/97 ----------------------------MTRRALLARAAAPLAPLALAMV
Rv0411c|M.tuberculosis_H37Rv ----------------------------MTRRALLARAAAPLAPLALAMV
MUL_2809|M.ulcerans_Agy99 ----------------------------MRRLPGLRRAASALAVLALVIT
MLBr_00303|M.leprae_Br4923 ---------------------------MMTRQFKIQRACS---VLATAMM
: :
Mflv_1232|M.gilvum_PYR-GCK VVANQVTITPERQQKYDLSEPYSIGEGVIVTRADDDSITSLDDLR--GRR
Mvan_5576|M.vanbaalenii_PYR-1 VVANQVTITPERQQKYDLSEPYSIGEGVIVTRADDDSITSLADLR--GKR
MSMEG_6307|M.smegmatis_MC2_155 IIANEVTITDERKAKYDLSEPYSVGEGVIVTRADDNSITSLDDLK--GKV
MAB_0277c|M.abscessus_ATCC_199 VVANQVTITPARKAKYDISTPYAVGEGVIVTRADDNSIHSLSDVR--GKV
MMAR_0061|M.marinum_M VGSASVKLTEARRRTVAFTNGYDFGYYSLVVPPG-SAITKFSDLAPGQRI
MAV_0059|M.avium_104 VGSSSIKATDERRQTVAFTNGYDFGYYALVVPPG-SAIKEFTDLAAGQRI
TH_0515|M.thermoresistible__bu VGSSSITTTEQRRRTVEFTNGYDFGYFSLVVPSG-SAITGFGDLVPGQRI
Mb0419c|M.bovis_AF2122/97 LASCGHSETLGVEATPTLPLPTPVGMEIMPPQPPLPPDSSSQDCDPTASL
Rv0411c|M.tuberculosis_H37Rv LASCGHSETLGVEATPTLPLPTPVGMEIMPPQPPLPPDSSSQDCDPTASL
MUL_2809|M.ulcerans_Agy99 LDGCGHSEPLKVESTPTLPLPTPVGMEVMPPEPPLPADSSSQDCDPTASL
MLBr_00303|M.leprae_Br4923 LAACGHAELLTVASMPTLPPPTPVGMEQVPPELLLPQDNTEEDCDPTASL
: . :. .* : *
Mflv_1232|M.gilvum_PYR-GCK AAQSTTSNWAQVARDAGAQIESVEGLTQAMALLSQGRVDVVVNDSLSIYA
Mvan_5576|M.vanbaalenii_PYR-1 AAQSTTSNWAGVARDAGAQIESVEGLTQAMALLSQGRVDVVVNDSLSIYA
MSMEG_6307|M.smegmatis_MC2_155 AAENATSNWSEIARQAGARVEAVEGFTQAIKLLNQGRVDVVVNDSIAVYA
MAB_0277c|M.abscessus_ATCC_199 AAENATSNWSQIARDAGARVEAVEGFTQSITLLSQGRVDVVINDSIAVYA
MMAR_0061|M.marinum_M GVVQGTVEESYVVNSLHLQPVKYPDFATVYGNLKTHQIDAWVAPGNQAEA
MAV_0059|M.avium_104 GVVQGTVEESYVVDTLHLQPVKYPDFATVYASLKTRQIDAWVAPAGQAAT
TH_0515|M.thermoresistible__bu GVVQGTVQESYVVDTLGLRPVIFPDYNTVYASLKTRQIDAWVAPSQQAEG
Mb0419c|M.bovis_AF2122/97 RPFATKAEADAAVADIRARGRLIVGLDIGSNLFSFR--DPITGEITGFDV
Rv0411c|M.tuberculosis_H37Rv RPFATKAEADAAVADIRARGRLIVGLDIGSNLFSFR--DPITGEITGFDV
MUL_2809|M.ulcerans_Agy99 RPFPTKAEADAAVADIRARGRLIVGLDIGSNLFSFR--DPITGEITGFDV
MLBr_00303|M.leprae_Br4923 RPFPTKAEADAAVAEIRARGRLIVGLEIGSNLFSFR--DPITGEITGFDI
. : . : . :. *
Mflv_1232|M.gilvum_PYR-GCK YLAETGDTAVKIAGTTGEKSEQGFAARK-NSGLLPDLNTAVEELKADGTM
Mvan_5576|M.vanbaalenii_PYR-1 YLAETNDTAVKIAGATGEKSEQGFAARK-NSGLLPDLNTALEELKADGTL
MSMEG_6307|M.smegmatis_MC2_155 YLAETGDTSVKIAGEVGEKSEQGFAARK-DSGYLPELNRALDELRADGTL
MAB_0277c|M.abscessus_ATCC_199 YLASTGDPAVKIAGATGQRSEQGFAARK-NSGLLPDLDKALDQLAADGTL
MMAR_0061|M.marinum_M AIRP-GDPAVLVANTFGAGDFVAYVVARDNRPLLDALNSGLDAVIADGTW
MAV_0059|M.avium_104 TIQP-GDPAVVVANTISPGNFVAYAVAKDNKPLVDALNSGLDAVIADGTW
TH_0515|M.thermoresistible__bu TVRP-GDPAEIIENTFSLDNFIAYAVAPGNQPLIDALNAGLDAVIADGTW
Mb0419c|M.bovis_AF2122/97 DIAG-----EVARDIFGVPSHVEYRILSAAERVTALQKSQVDIVVKTMSI
Rv0411c|M.tuberculosis_H37Rv DIAG-----EVARDIFGVPSHVEYRILSAAERVTALQKSQVDIVVKTMSI
MUL_2809|M.ulcerans_Agy99 DIAS-----EVARDIFGGPTHVEYRILSAAERITALQSSQVDVVVKTMTI
MLBr_00303|M.leprae_Br4923 DIAG-----EVARDIFGTPSHMEYRILSTAARITALQKSEVDIIVKTMTI
: . . : . :: : :
Mflv_1232|M.gilvum_PYR-GCK AEISQKYLKTNASGG-------------------------QDTPQAQP--
Mvan_5576|M.vanbaalenii_PYR-1 AEISQKYLKTNASGG-------------------------QDAPQTQP--
MSMEG_6307|M.smegmatis_MC2_155 AQISQKYLKADATGA-------------------------ANGDGAPPAV
MAB_0277c|M.abscessus_ATCC_199 TRISQKYLKTDASGTTKP----------------------QAGKGDSGST
MMAR_0061|M.marinum_M SQLYSKWVPRTLPPGWKPGSRAVAAPTLPDFAAIAAA--HRHKPVGPAAP
MAV_0059|M.avium_104 SNLYSQWVPRTLPPGWKPGSQAVAPPKLPDFAAIAAS--QHHKAVGPAAP
TH_0515|M.thermoresistible__bu ARLYSEWVPRALPPGWKPGSKAAPPPQLPDFDAIAAERASQQPAAGPGVR
Mb0419c|M.bovis_AF2122/97 TCERRKLVN-----------------------------------------
Rv0411c|M.tuberculosis_H37Rv TCERRKLVN-----------------------------------------
MUL_2809|M.ulcerans_Agy99 TCERRKLVN-----------------------------------------
MLBr_00303|M.leprae_Br4923 TCKRRKLVN-----------------------------------------
: : :
Mflv_1232|M.gilvum_PYR-GCK RSTLQLVLDNLWP----------LAKAALTMTIPLTIISFVIGLVIALGV
Mvan_5576|M.vanbaalenii_PYR-1 RSALRLVLDNLWP----------LARAALTMTIPLTIISFVIGLVIALGV
MSMEG_6307|M.smegmatis_MC2_155 RSTWQLIGDNLWP----------LAKAAVTMTIPLTIISFAIGLVIALGV
MAB_0277c|M.abscessus_ATCC_199 RSDTQLVLDNLWP----------MARAMITVTLPLTAISFVIGLVIALGV
MMAR_0061|M.marinum_M KSALTQLRDSFFDWDMYRQAIPTLFKTGLPNTLILTLSASIIGLALGMAL
MAV_0059|M.avium_104 KSTLAQLRDSFFDWDMYRQAIPTLLRTGLPNTLILTFSASVIGLVLGMVL
TH_0515|M.thermoresistible__bu KSTLAQLADSFLDWELYRQAIPDLLTTGLPNTLLLTVGAGVIGLVLGMVL
Mb0419c|M.bovis_AF2122/97 -----------------------------FSTVYLDANQRILAPRDSPIT
Rv0411c|M.tuberculosis_H37Rv -----------------------------FSTVYLDANQRILAPRDSPIT
MUL_2809|M.ulcerans_Agy99 -----------------------------FSTVYLDANQRILTSRDSPIF
MLBr_00303|M.leprae_Br4923 -----------------------------FSTVYLDAKQRILAPRDSPIT
*: * : .
Mflv_1232|M.gilvum_PYR-GCK ALARLSSNVVLSNLARFYISVIRGTPLLVQLFIVFFALPEFG---VKIDP
Mvan_5576|M.vanbaalenii_PYR-1 ALARLSSNVVLTNLARFYISVIRGTPLLVQLFIVFFALPEFG---VRIDP
MSMEG_6307|M.smegmatis_MC2_155 ALARLSSNAVLSNVARLYISIIRGTPLLVQLFLVFYALPEFG---VRIDP
MAB_0277c|M.abscessus_ATCC_199 ALARMSGHRVVTGLARIYISVIRGTPLLLQLFLIFFALPEFG---VKISP
MMAR_0061|M.marinum_M AIAGISHQRWLRWPARVYTDIFRGLPEVVIILIIGLGVGPIVGGLTNNNP
MAV_0059|M.avium_104 AVAGISHARWLRWPARVYTDVFRGLPEVVIILLIGLGIGPIVGGLTNNNP
TH_0515|M.thermoresistible__bu AIAGISRSRLLRWPARVYTDIFRGLPEVVIILLIGLGLGPVVGGLTGYNP
Mb0419c|M.bovis_AF2122/97 KVSDLSGKRVCVARGTTSLRRIR-------------------------EI
Rv0411c|M.tuberculosis_H37Rv KVSDLSGKRVCVARGTTSLRRIR-------------------------EI
MUL_2809|M.ulcerans_Agy99 KVSDLSGKRVCVARGTTSLRRIR-------------------------EI
MLBr_00303|M.leprae_Br4923 KPSDLSNKRVCMTRGTTSLHQVS-------------------------EV
: :* . . .
Mflv_1232|M.gilvum_PYR-GCK FPAAVIAFSLNVGGYAAEIIRSAIQSIPKGQWEAAETIGLNYVGTLRRII
Mvan_5576|M.vanbaalenii_PYR-1 FPAAVIAFSLNVGGYAAEIIRSAIQSIPKGQWEAAETIGLNYAGTLRRII
MSMEG_6307|M.smegmatis_MC2_155 FPAAVIAFSLNVGGYAAEIIRAAILSIPKGQWEAAETIGLTYGKSLQRII
MAB_0277c|M.abscessus_ATCC_199 FPAAVIAFSLNVGGYAAEIIRSSILSIPRGQWEAASSLGMSYSTTLWRIV
MMAR_0061|M.marinum_M YPLGIAALGLMAGAYVGEILRSGIQSVDPGQLEASRALGFSYSAAMRLVV
MAV_0059|M.avium_104 YPLGIAALGMMAAAYIGEILRSGIQSVDPGQLEASRALGFSYATAMRLVV
TH_0515|M.thermoresistible__bu FPLGILALGLMAAAYVGEILRSGIQSVEPGQLEAARALGFSYPAAMRLVV
Mb0419c|M.bovis_AF2122/97 APPPVIVSVVNWADCLVALQQREIDAVST---DDTILAGLVEEDPYLHIV
Rv0411c|M.tuberculosis_H37Rv APPPVIVSVVNWADCLVALQQREIDAVST---DDTILAGLVEEDPYLHIV
MUL_2809|M.ulcerans_Agy99 APPPVIVSVVNWADCLVALQQREIDAVST---DDTILAGLVEEDPYLHIV
MLBr_00303|M.leprae_Br4923 VPPPIIVSAENWADCLVAMQQREIDAIST---DDAILAGLVEEDPYLHIV
* : . . : : * :: : : *: . ::
Mflv_1232|M.gilvum_PYR-GCK LPQATRVAVPPLSNTLISLVKDTSLASTILVTELLR---QAQIIAAPTFE
Mvan_5576|M.vanbaalenii_PYR-1 LPQATRVAVPPLSNTLISLVKDTSLASTILVTELLR---QAQIIAAPTFE
MSMEG_6307|M.smegmatis_MC2_155 LPQAARVAVPPLSNTLISLVKDTSLASTILVTELLR---QAQIAAAPTFE
MAB_0277c|M.abscessus_ATCC_199 IPQASRVAVPPLSNTLISLVKDTSLASAILVTDVMR---TAQVAAAPTFQ
MMAR_0061|M.marinum_M VPQGVRRVLPALVNQFIALLKASALVYFLGLIAQQRELFQVGRDLNAQTG
MAV_0059|M.avium_104 VPQGVRRVLPALVNQFIALLKASALVYFLGLVADQRELFQVGRDLNAETG
TH_0515|M.thermoresistible__bu VPQGVRRVLPALMNQFISLLKASSLVYFLGLVADQRELFQVGRDLNAQTG
Mb0419c|M.bovis_AF2122/97 GPDMADQPYGVGIN-----LDNTGLVRFVN-------------------G
Rv0411c|M.tuberculosis_H37Rv GPDMADQPYGVGIN-----LDNTGLVRFVN-------------------G
MUL_2809|M.ulcerans_Agy99 GPNMANQPYGIGIN-----LDNTGLVRFVN-------------------G
MLBr_00303|M.leprae_Br4923 GPNMATQPYGIGIN-----LNNTTLVRFVN-------------------G
*: * :. : *. :
Mflv_1232|M.gilvum_PYR-GCK FFALYGTAAVYYWVICLVLSFGQARFEHRLERYVAR--------------
Mvan_5576|M.vanbaalenii_PYR-1 FFALYGTAAVYYWVICLVLSFGQARFERRLERYVAK--------------
MSMEG_6307|M.smegmatis_MC2_155 FFALYGTAAVYYWVICLVLSFGQSRIERRLERYVAR--------------
MAB_0277c|M.abscessus_ATCC_199 FFTLYVTAGVYYWIVCMAMSAVQDRLEQRLEKSVAR--------------
MMAR_0061|M.marinum_M NLSPLVAAGIFYLMLTVPLTHLVNYIDARLRRGRRRLDPEDPPGVVSSTI
MAV_0059|M.avium_104 NLSPLVAAGACYLILTVPLTHLVNMIDARLRRGRAAVDPEEPAEMLT--F
TH_0515|M.thermoresistible__bu SLSPLVAAGLFYLALTIPLTHLVNYVDNRLRRGRK-PAAEDPLEPAT---
Mb0419c|M.bovis_AF2122/97 TLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD---------------
Rv0411c|M.tuberculosis_H37Rv TLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD---------------
MUL_2809|M.ulcerans_Agy99 TLERIRNDGTWNTLYRKWLSVLGPAPAPPRPRYVD---------------
MLBr_00303|M.leprae_Br4923 TLERIRRDGTWITLYRKWLTVLGPAPAPPTPRYVD---------------
: . :: :
Mflv_1232|M.gilvum_PYR-GCK -----
Mvan_5576|M.vanbaalenii_PYR-1 -----
MSMEG_6307|M.smegmatis_MC2_155 -----
MAB_0277c|M.abscessus_ATCC_199 -----
MMAR_0061|M.marinum_M GEEMT
MAV_0059|M.avium_104 GQEIT
TH_0515|M.thermoresistible__bu AQEMI
Mb0419c|M.bovis_AF2122/97 -----
Rv0411c|M.tuberculosis_H37Rv -----
MUL_2809|M.ulcerans_Agy99 -----
MLBr_00303|M.leprae_Br4923 -----