For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGVSGAVPVPQRMCRQGGRPRRVGPVIDPKVLRENPDAVRASQQARGEDPSLVDALAEADSARRAAISA ADNLRAEQKSASKAIGKASPDERPALLAAAKELSEKVKAAEADQAAAEAAYTAAHMAISNVIIDGVPAGG EDDFVVLDTVGEPRVIPDAKDHLELGEALGLIDMERGAKVSGSRFYFLTGFGALLQLGLFQLAVRTATEN GFTLMIPPVLVRPEIMRGTGFLGAHADEIYHLAEDDLYLVGTSEVPLAGYHSDEILDLSGGPLRYAGWSS CFRREAGSYGKDTRGIIRVHQFDKVEGFVYCRPEEAEAEHDRLLGWQRQLLAAIEVPYRVIDIAAGDLGS SAARKYDCEAWVPTQQTYRELTSTSNCTTFQARRLAVRYRDENGRPQTAATLNGTLATTRWLVAILENHQ QPDGSVRIPEALVPHVGTNVLRPKK
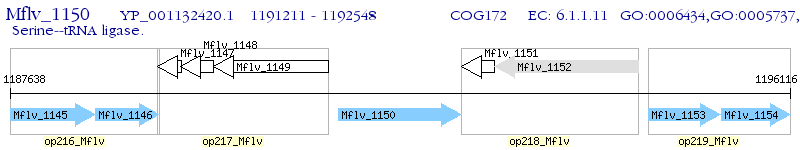
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1150 | - | - | 100% (445) | seryl-tRNA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3864c | serS | 0.0 | 79.62% (417) | seryl-tRNA synthetase |
| M. tuberculosis H37Rv | Rv3834c | serS | 0.0 | 79.62% (417) | seryl-tRNA synthetase |
| M. leprae Br4923 | MLBr_00082 | serS | 0.0 | 77.35% (415) | seryl-tRNA synthetase |
| M. abscessus ATCC 19977 | MAB_0153 | - | 0.0 | 80.62% (418) | seryl-tRNA synthetase |
| M. marinum M | MMAR_5386 | serS | 0.0 | 79.90% (418) | seryl-tRNA synthetase SerS |
| M. avium 104 | MAV_0194 | serS | 0.0 | 81.43% (420) | seryl-tRNA synthetase |
| M. smegmatis MC2 155 | MSMEG_6413 | serS | 0.0 | 84.65% (417) | seryl-tRNA synthetase |
| M. thermoresistible (build 8) | TH_0892 | serS | 0.0 | 83.10% (420) | SERYL-TRNA SYNTHETASE SERS (SERINE--TRNA LIGASE) (SERRS) |
| M. ulcerans Agy99 | MUL_5006 | serS | 0.0 | 79.43% (418) | seryl-tRNA synthetase |
| M. vanbaalenii PYR-1 | Mvan_5659 | - | 0.0 | 89.95% (418) | seryl-tRNA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1150|M.gilvum_PYR-GCK MTGVSGAVPVPQRMCRQGGRPRRVGPVIDPKVLRENPDAVRASQQARGED
Mvan_5659|M.vanbaalenii_PYR-1 --------------------------MIDLKVLRENPDAVRASQLARGED
Mb3864c|M.bovis_AF2122/97 --------------------------MIDLKLLRENPDAVRRSQLSRGED
Rv3834c|M.tuberculosis_H37Rv --------------------------MIDLKLLRENPDAVRRSQLSRGED
MMAR_5386|M.marinum_M --------------------------MIDLKLLREDPDVVRRSQLSRGED
MUL_5006|M.ulcerans_Agy99 --------------------------MIDLKLLREDPDVVRRSQLSRGED
MLBr_00082|M.leprae_Br4923 --------------------------MIDLQLLREDPDVVRRSQLSRGED
MAV_0194|M.avium_104 --------------------------MIDLKLLREDPDAVRRSQQSRGED
MSMEG_6413|M.smegmatis_MC2_155 --------------------------MIDLRLLRENPDIVRASQRARGED
TH_0892|M.thermoresistible__bu -----------------------VAAVIDLKLLRENPDVVRASQRARGED
MAB_0153|M.abscessus_ATCC_1997 --------------------------MIDLKFLRENPDAVRASQRLRGED
:** :.***:** ** ** ****
Mflv_1150|M.gilvum_PYR-GCK PSLVDALAEADSARRAAISAADNLRAEQKSASKAIGKASPDERPALLAAA
Mvan_5659|M.vanbaalenii_PYR-1 PGLVDALADADTARRAAISAADNLRAEQKSASRLVGKASPEERPALLAAA
Mb3864c|M.bovis_AF2122/97 PALVDALLTADAARRAVISTADSLRAEQKAASKSVGGASPEERPPLLRRA
Rv3834c|M.tuberculosis_H37Rv PALVDALLTADAARRAVISTADSLRAEQKAASKSVGGASPEERPPLLRRA
MMAR_5386|M.marinum_M PALVDALLTADTARRTAISTADSLRAEQKAASKGVGGASPEERPALLQRA
MUL_5006|M.ulcerans_Agy99 PALVDALLTADTARRTAISTADSLRAEQKAASKSVGGASPEERPALLQRA
MLBr_00082|M.leprae_Br4923 PALVDALLTADTARRAAISAADSLRAEQKATSKSLGAASAEDRPALLERA
MAV_0194|M.avium_104 PSLVDALLAADAARRAAISTADTLRAEQKSASKSVGAASPEQRPALLARA
MSMEG_6413|M.smegmatis_MC2_155 PALVDALLAADTARRSAVSAADNLRAEQKAASKLVGKASPDERPALLQRA
TH_0892|M.thermoresistible__bu PALVDALLAADEARRAAVSAADNLRAEQKAASRSVGKASPDERPALLERA
MAB_0153|M.abscessus_ATCC_1997 PALVEVLLDADTARRAAISTADTLRAEQKAASKKVGKATPEERPALLAAA
*.**:.* ** ***:.:*:**.******::*: :* *:.::**.** *
Mflv_1150|M.gilvum_PYR-GCK KELSEKVKAAEADQAAAEAAYTAAHMAISNVIIDGVPAGGEDDFVVLDTV
Mvan_5659|M.vanbaalenii_PYR-1 KELADKVRAAESAQAEAEKAYSAAHMAISNVIIDGVPAGGEDDFVVLDTV
Mb3864c|M.bovis_AF2122/97 KELAEQVKAAEADEVEAEAAFTAAHLAISNVIVDGVPAGGEDDYAVLDVV
Rv3834c|M.tuberculosis_H37Rv KELAEQVKAAEADEVEAEAAFTAAHLAISNVIVDGVPAGGEDDYAVLDVV
MMAR_5386|M.marinum_M KDLAEQVKAAEATQSETEAAFTAAHMAIPNVILDGVPAGGEDDYAVLEVV
MUL_5006|M.ulcerans_Agy99 KDLAEQVKAAEATQSETETAFTAAHMAIPNVILDGVPAGGEDDYAVLDVV
MLBr_00082|M.leprae_Br4923 KDLAEQVKAAETTQAETAAAFAAAYMAISNVVLDGVPTGGKDDYAVLDIV
MAV_0194|M.avium_104 KELAEQVKAAETAQAEAEAAFTAAHLAISNVVIDGVPAGGEDDFAVLDVV
MSMEG_6413|M.smegmatis_MC2_155 KDLAEQVKAAEAAQAEAEQAFTAAHMAISNVIFEGVPAGGEDDFVVLDTV
TH_0892|M.thermoresistible__bu KELAEQVKAAEARQAEAEQALTEAHMAISNIVIDGVPPGGEDDFVVLDTV
MAB_0153|M.abscessus_ATCC_1997 SELAAQVKSAEAEQVVTEAAFTEAHKAISNVVIDGVPAGGEQDFALRELV
.:*: :*::**: : : * : *: **.*::.:***.**::*:.: : *
Mflv_1150|M.gilvum_PYR-GCK GEPRVIPDAKDHLELGEALGLIDMERGAKVSGSRFYFLTGFGALLQLGLF
Mvan_5659|M.vanbaalenii_PYR-1 GEPTALTDPKDHLELGESLGLIDMERGAKVSGSRFYFLTGFGALLQLGLF
Mb3864c|M.bovis_AF2122/97 GEPSYLENPKDHLELGESLGLIDMQRGAKVSGSRFYFLTGRGALLQLGLL
Rv3834c|M.tuberculosis_H37Rv GEPSYLENPKDHLELGESLGLIDMQRGAKVSGSRFYFLTGRGALLQLGLL
MMAR_5386|M.marinum_M GEPRAIDNPKDHLELGESLGLIDMARGAKVAGSRFYFLTGQGALLQLGLL
MUL_5006|M.ulcerans_Agy99 GEPRALDNPKDHLELGESLGLIDMARGAKVAGSRFYFLTGQGALLQLGLL
MLBr_00082|M.leprae_Br4923 GDPPPLSNPKDHLELGEALGLIDMQRGAKVSGSRFYFLTGRGALLQLGLL
MAV_0194|M.avium_104 GEPTALENPRDHLELGESLGLIDMARGAKVSGSRFYFLTGKGALLQLGLL
MSMEG_6413|M.smegmatis_MC2_155 GEPRAIENPKDHLELGESLGLIDMERGAKVSGSRFYFLTGAGALLQLGLL
TH_0892|M.thermoresistible__bu GEPTPIDNPRDHLELGESLGLIDVERGAKVSGSRFYFLTGAGALLQLGLL
MAB_0153|M.abscessus_ATCC_1997 GEPHPIENPKDHVELGESLGLFDMERGAKVSGARFYFLTGYGALLQLGLL
*:* : :.:**:****:***:*: *****:*:******* ********:
Mflv_1150|M.gilvum_PYR-GCK QLAVRTATENGFTLMIPPVLVRPEIMRGTGFLGAHADEIYHLAEDDLYLV
Mvan_5659|M.vanbaalenii_PYR-1 QLAVRTATENGFTLVIPPVLVRPEIMSGTGFLGAHADEVYRVEADDLYLV
Mb3864c|M.bovis_AF2122/97 QLALKLAVDNGFVPTIPPVLVRPEVMVGTGFLGAHAEEVYRVEGDGLYLV
Rv3834c|M.tuberculosis_H37Rv QLALKLAVDNGFVPTIPPVLVRPEVMVGTGFLGAHAEEVYRVEGDGLYLV
MMAR_5386|M.marinum_M QLALRLAVDNGFIPMIPPVLVRPEVMAGTGFLGAHADEVYRLEADDLYLV
MUL_5006|M.ulcerans_Agy99 QLALRLAVDNGFIPMIPPVLVRPEVMAGTGFLGAHADEVYRLEADDLYLV
MLBr_00082|M.leprae_Br4923 QLALRLAVENDFIPMIPPVLVRPEVMSGTGFLGAHAEEVYR--VDDLYLV
MAV_0194|M.avium_104 QLALRLAVENGFVPMIPPVLVRPEVMAGTGFLGAHAEEVYRIEADDLYLV
MSMEG_6413|M.smegmatis_MC2_155 QLATQVAVQNGFTLMIPPVLVRPEVMRGTGFLGAHADEVYRLEADDMYLV
TH_0892|M.thermoresistible__bu QLAMRVATQNGFTPMIPPVLVRPQIMSGTGFLGAHAAEVYRIESDDLYLV
MAB_0153|M.abscessus_ATCC_1997 QLAVQTAVANGFTLLIPPVLVKPEIMGGTGFLGAHADEVYHLEEDDLYLV
*** : *. *.* ******:*::* ********* *:*: *.:***
Mflv_1150|M.gilvum_PYR-GCK GTSEVPLAGYHSDEILDLSGGPLRYAGWSSCFRREAGSYGKDTRGIIRVH
Mvan_5659|M.vanbaalenii_PYR-1 GTSEVPLAGYHSGEILDLSEGPKRYAGWSSCFRREAGSYGKDTRGIIRVH
Mb3864c|M.bovis_AF2122/97 GTSEVPLAGYHSGEILDLSRGPLRYAGWSSCFRREAGSHGKDTRGIIRVH
Rv3834c|M.tuberculosis_H37Rv GTSEVPLAGYHSGEILDLSRGPLRYAGWSSCFRREAGSHGKDTRGIIRVH
MMAR_5386|M.marinum_M GTSEVPMAGYHADEIVDLSEGPLRYAGWSSCFRREAGSYGKDTRGIIRVH
MUL_5006|M.ulcerans_Agy99 GTSEVPMAGYHADEIVDLSEGPLRYAGWSSCFRREAGSYGKDTRGIIRVH
MLBr_00082|M.leprae_Br4923 GTSEVPMAGYHSDEILDLSVGPLRYAGWSSCFRREAGSHGKDTRGIIRVH
MAV_0194|M.avium_104 GTSEVPLAGYHADEILDLSAGPLRYAGWSSCFRREAGSYGKDTRGVIRVH
MSMEG_6413|M.smegmatis_MC2_155 GTSEVPLAGYHADEILDLSAGPRRYAGWSSCFRREAGSYGKDTRGIIRVH
TH_0892|M.thermoresistible__bu GTSEVPLAGYHADEILDLSNGPLRYAGWSSCFRREAGSYGKDTRGIIRVH
MAB_0153|M.abscessus_ATCC_1997 GTSEVPIAGYHSGEILDLNNGPLRYAGWSSCFRREAGSYGKDTRGIIRVH
******:****:.**:**. ** ***************:******:****
Mflv_1150|M.gilvum_PYR-GCK QFDKVEGFVYCRPEEAEAEHDRLLGWQRQLLAAIEVPYRVIDIAAGDLGS
Mvan_5659|M.vanbaalenii_PYR-1 QFDKVEGFVYCRPEEAEAEHDRLLGWQRQMLGLIEVPYRVIDIAAGDLGS
Mb3864c|M.bovis_AF2122/97 QFDKVEGFVYCTPADAEHEHERLLGWQRQMLARIEVPYRVIDVAAGDLGS
Rv3834c|M.tuberculosis_H37Rv QFDKVEGFVYCTPADAEHEHERLLGWQRQMLARIEVPYRVIDVAAGDLGS
MMAR_5386|M.marinum_M QFDKVEGFVYCTPADAEAEHQRLLAWQREMLAQIEVPYRVIDVAAGDLGA
MUL_5006|M.ulcerans_Agy99 QFDKVEGFVYCAPADAEAEHQRLLAWQREMLAQIEVPYRVIDVAAGDLGA
MLBr_00082|M.leprae_Br4923 QFDKVEGFVYCAPTDAEVEHQRLLGWQREMLARIEVPYRVIDVAAGDLGS
MAV_0194|M.avium_104 QFDKVEGFVYCTPADAEAEHQRLLGWQREMLARIEVPYRVIDVAAGDLGS
MSMEG_6413|M.smegmatis_MC2_155 QFDKVEGFIYCKPEDAEAEHQRLLGWQREMLAAIEVPYRVIDVAAGDLGS
TH_0892|M.thermoresistible__bu QFDKVEAFVYCRPEEAEAEHDRLLAWQKKMLAHIEVPYRVIDVAAGDLGA
MAB_0153|M.abscessus_ATCC_1997 QFDKVEAFVYCKPEDAEAEHEKILGWERQMLGHIEVPYRVIDVAAGDLGS
******.*:** * :** **:::*.*::::*. *********:******:
Mflv_1150|M.gilvum_PYR-GCK SAARKYDCEAWVPTQQTYRELTSTSNCTTFQARRLAVRYR---DENGRPQ
Mvan_5659|M.vanbaalenii_PYR-1 SAARKFDCEAWVPTQQTYRELTSTSNCTTFQARRLGVRYR---DENGKPQ
Mb3864c|M.bovis_AF2122/97 SAARKFDCEAWIPTQGAYRELTSTSNCTTFQARRLATRYR---DASGKPQ
Rv3834c|M.tuberculosis_H37Rv SAARKFDCEAWIPTQGAYRELTSTSNCTTFQARRLATRYR---DASGKPQ
MMAR_5386|M.marinum_M SAARKFDCEAWVPTQGTYRELTSTSNCTTFQARRLATRYR---DANGKPQ
MUL_5006|M.ulcerans_Agy99 SAARKFDCEAWVPTQGTYRELTSTSNCTTFQARRLATRYR---DANGKPQ
MLBr_00082|M.leprae_Br4923 SAARKFDCEAWVPTQGSYCELTSTSNCTTFQARRLATRYR---DASGKPQ
MAV_0194|M.avium_104 SAARKFDCEAWVPTQQTYRELTSTSNCTTFQARRLSTRYRDGGDAKGKPQ
MSMEG_6413|M.smegmatis_MC2_155 SAARKYDCEAWVPTQQTYRELTSTSNCTTFQARRLSTRYR---DDNGKPQ
TH_0892|M.thermoresistible__bu SAARKYDCEAWLPSQQAYRELTSTSNCTTFQARRLATRYR---DENGRPQ
MAB_0153|M.abscessus_ATCC_1997 SAARKFDCEAWVPTQRAYRELTSTSNCTTFQARRLAVRYR---DESGKPQ
*****:*****:*:* :* ****************..*** * .*:**
Mflv_1150|M.gilvum_PYR-GCK TAATLNGTLATTRWLVAILENHQQPDGSVRIPEALVPHVGTNVLRPKK--
Mvan_5659|M.vanbaalenii_PYR-1 TAATLNGTLATTRWLVAILENHQQPDGSVRIPEALVPYVGTEVLTPKN--
Mb3864c|M.bovis_AF2122/97 IAATLNGTLATTRWLVAILENHQRPDGSVRVPDALVPFVGVEVLEPVA--
Rv3834c|M.tuberculosis_H37Rv IAATLNGTLATTRWLVAILENHQRPDGSVRVPDALVPFVGVEVLEPVA--
MMAR_5386|M.marinum_M IAATLNGTLGTTRWLVSILENHQQPDGSVRVPAALVGFVGTEVLEPKG--
MUL_5006|M.ulcerans_Agy99 IAATLNGTLGTTRWLVSILENHQQPDGSARVPAALVGFVGTEVLEPKG--
MLBr_00082|M.leprae_Br4923 IVATLNGTLGTTRWLVAILENHQRPDGSVRVPPALVPFVGTELLESPR--
MAV_0194|M.avium_104 IAATLNGTLGTTRWLVAILENHQRPDGSVRVPEALVPFVGTELLEPVGPR
MSMEG_6413|M.smegmatis_MC2_155 IAATLNGTLATTRWLVAILENHQQPDGSVRVPAALVPYVRTEVLEP----
TH_0892|M.thermoresistible__bu IAATLNGTLATTRWLVAILENHQRPDGSVRVPEALRPYVGVEVLEP----
MAB_0153|M.abscessus_ATCC_1997 IAATLNGTLATTRWLVAILENHQQPDGSVRVPTALVPFVGTEVLEPK---
.*******.******:******:****.*:* ** .* .::* .
Mflv_1150|M.gilvum_PYR-GCK -
Mvan_5659|M.vanbaalenii_PYR-1 -
Mb3864c|M.bovis_AF2122/97 -
Rv3834c|M.tuberculosis_H37Rv -
MMAR_5386|M.marinum_M -
MUL_5006|M.ulcerans_Agy99 -
MLBr_00082|M.leprae_Br4923 -
MAV_0194|M.avium_104 G
MSMEG_6413|M.smegmatis_MC2_155 -
TH_0892|M.thermoresistible__bu -
MAB_0153|M.abscessus_ATCC_1997 -