For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDYGYFLQITTRWMDNDVYGHVNNVTYYSYFDTVANHFLIREGGLDIHTSPVIALVVESKCSYLAPVAY PDDLRAGLRVDKLTNRSVTYGVAIFAGDGDDPVAHGYFVHVFVDREHRRAVPIPETVRAALERLRGQ
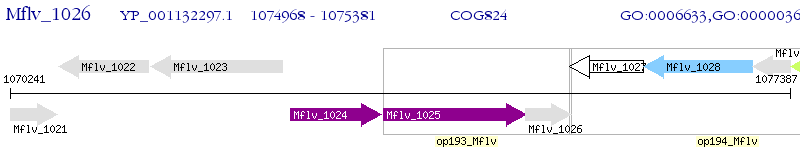
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1026 | - | - | 100% (137) | thioesterase superfamily protein |
| M. gilvum PYR-GCK | Mflv_3302 | - | 8e-06 | 27.86% (140) | thioesterase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0168 | - | 6e-28 | 43.18% (132) | hypothetical protein Mb0168 |
| M. tuberculosis H37Rv | Rv0163 | - | 6e-28 | 43.18% (132) | hypothetical protein Rv0163 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0982c | - | 4e-32 | 49.28% (138) | putative acyl-CoA hydrolase/thioesterase |
| M. marinum M | MMAR_0406 | - | 1e-26 | 41.30% (138) | hypothetical protein MMAR_0406 |
| M. avium 104 | MAV_5021 | - | 6e-28 | 41.04% (134) | thioesterase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_0128 | - | 6e-27 | 44.62% (130) | thioesterase superfamily protein |
| M. thermoresistible (build 8) | TH_0158 | - | 2e-32 | 47.01% (134) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1056 | - | 3e-27 | 42.03% (138) | hypothetical protein MUL_1056 |
| M. vanbaalenii PYR-1 | Mvan_5810 | - | 6e-66 | 87.12% (132) | thioesterase superfamily protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0406|M.marinum_M MPAGPSPDELSSGDFPVLWPVLTRWADNDMFGHLNNAVYYQLFDSAINAW
MUL_1056|M.ulcerans_Agy99 MPAGPSPDELSSGDFPVLWPVLTRWADNDMFGHLNNAVYYQLFDTAINAW
MAV_5021|M.avium_104 ---------MTSNDFPVLWPVGTRWADNDMFGHLNNAVYYQLFDTAINGW
Mb0168|M.bovis_AF2122/97 MAALPAPEKLLRSDFPVLWPVGTRWADNDMFGHLNNAVYYQLFDTAINAW
Rv0163|M.tuberculosis_H37Rv MAALPAPEKLLRSDFPVLWPVGTRWADNDMFGHLNNAVYYQLFDTAINAW
MSMEG_0128|M.smegmatis_MC2_155 -MTRRDEPTPTGREFPVHWPVGTRWTDNDMFGHLNNAVYYELFDTAINAW
TH_0158|M.thermoresistible__bu -MTS-DERTLTGRDFKVHWPVQTRWADNDIFGHLNNAAYYALFDTAINAW
MAB_0982c|M.abscessus_ATCC_199 ---MTKPELPTKADFPVHRRHQTRWADNDVYGHVNNVVYYAWFDTAVNGW
Mflv_1026|M.gilvum_PYR-GCK -----------MSDYGYFLQITTRWMDNDVYGHVNNVTYYSYFDTVANHF
Mvan_5810|M.vanbaalenii_PYR-1 -----MPAQHRPEDYGYFVPITTRWMDNDVYGHVNNVTYYSYFDTVANHF
:: *** ***::**:**..** **:. * :
MMAR_0406|M.marinum_M INTRTGLDPLTTAALGIVAESGCRYFSELHFPQELVVGLAVTRLGRSSVT
MUL_1056|M.ulcerans_Agy99 INTRTGLDPLTTAALGIVAESGCRYFSELHFPQELVVGLAVTRLGRSSVT
MAV_5021|M.avium_104 INTSTGLDPLTTPALGIVAESGCRYLSELHFPEGLQVGLAVTRLGRSSVT
Mb0168|M.bovis_AF2122/97 INTSTGVDPLAMPVLGIVAESGCRYFSELRFPESLMVGLAVTRLGRSSVT
Rv0163|M.tuberculosis_H37Rv INTSTGVDPLAMPVLGIVAESGCRYFSELRFPESLMVGLAVTRLGRSSVT
MSMEG_0128|M.smegmatis_MC2_155 IQTSCDVDPVTAPWLGVVAESGCRYFTELKFPGRLVVGLAVSRLGTSSVT
TH_0158|M.thermoresistible__bu INTTVGIDPLAAPWLGVVAESGCRYYAELRFPDPLVVGLKVTRLGNSSVT
MAB_0982c|M.abscessus_ATCC_199 LMEAAGCDIRDLPAIGVVAETSCKYLAEVSFPDELSIGLALEHSGRSSVI
Mflv_1026|M.gilvum_PYR-GCK LIREGGLDIHTSPVIALVVESKCSYLAPVAYPDDLRAGLRVDKLTNRSVT
Mvan_5810|M.vanbaalenii_PYR-1 LIREGGLDIHTAPVIGLVVESKCSYLAPVAYPDELRAGLRVDKLTNRSVT
: . * . :.:*.*: * * : : :* * ** : : **
MMAR_0406|M.marinum_M YRLAVFNSSPGAGEGPAGPGPIAALGHWVHVYVDRVSRKPVPIPETIRTL
MUL_1056|M.ulcerans_Agy99 YRLAVFNCSPGAGEGPAGQGPIAALGHWVHVYVDRVSRKPVPIPETIRTL
MAV_5021|M.avium_104 YRLGVFRS-----AAPGSAEPITALGHWVHVYVDRDTRKPVPIPDPIRTL
Mb0168|M.bovis_AF2122/97 YRLGVFKE-------PDDAGVITALGHWVHVYVDRTSRRPVPIPEAIRSL
Rv0163|M.tuberculosis_H37Rv YRLGVFKE-------PDDAGVITALGHWVHVYVDRTSRRPVPIPEAIRSL
MSMEG_0128|M.smegmatis_MC2_155 YRLALFAE-------DAADEPVAAVGHWVHVYVDRHTRRPVPIPDAIRDL
TH_0158|M.thermoresistible__bu YRLALFTG-------DGT-APVAAVGHWVHVYVDRSSRRPVPIPDAIREL
MAB_0982c|M.abscessus_ATCC_199 YKLAVFKVVD-----GVVEDTPRALGRFVHVYVDSDARTPVPIPEQVATA
Mflv_1026|M.gilvum_PYR-GCK YGVAIFAG---------DGDDPVAHGYFVHVFVDREHRRAVPIPETVRAA
Mvan_5810|M.vanbaalenii_PYR-1 YGVGIFSG---------TSDEAVAHGYFVHVFVDRERRTAVPIPEAIRAA
* :.:* * * :***:** * .****: :
MMAR_0406|M.marinum_M LATACVN-----
MUL_1056|M.ulcerans_Agy99 LATACVN-----
MAV_5021|M.avium_104 LAGAAVGDAAPK
Mb0168|M.bovis_AF2122/97 LSTACVSG----
Rv0163|M.tuberculosis_H37Rv LSTACVSG----
MSMEG_0128|M.smegmatis_MC2_155 LASAAGR-----
TH_0158|M.thermoresistible__bu LETAR-------
MAB_0982c|M.abscessus_ATCC_199 LEQLR-------
Mflv_1026|M.gilvum_PYR-GCK LERLRGQ-----
Mvan_5810|M.vanbaalenii_PYR-1 LTSLVVG-----
*