For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQTSQRELGDTDSPIEDALENAFNRMLDEGTQRLHRSWHEVLVTGFFGGTEVAMGVLAYLSVLQATGNQL LAGIAFSIGFLALLLGRSELFTEGFLIPVTTVAAKRAGIGQLAKLWGGTLIANLAGGWLIMWLVMTAYPK LHEKTTTSAEHFVNAPFNAETVALALLGGMVITLMTRMQHGTDSVPGKIAAAVAGAFLLAGLQMFHSILD SLLIFGALTAGDAPFGYLDWLSWFGYTLVLNVVGGLALVTLLRLIRSKDRLKEERRDAESPTAE*
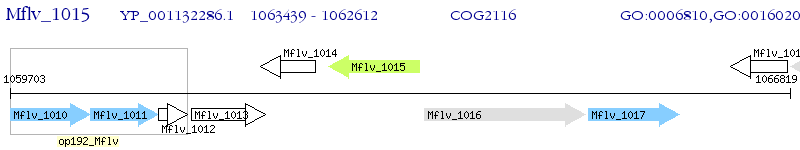
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1015 | - | - | 100% (275) | formate/nitrate transporter |
| M. gilvum PYR-GCK | Mflv_1996 | - | 2e-13 | 27.78% (270) | formate/nitrite transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2490 | - | 1e-05 | 24.53% (265) | formate/nitrite transporter superfamily protein |
| M. smegmatis MC2 155 | MSMEG_4318 | - | 1e-123 | 78.23% (271) | hypothetical protein MSMEG_4318 |
| M. thermoresistible (build 8) | TH_1321 | - | 1e-125 | 80.07% (271) | hypothetical protein MSMEG_4318 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4123 | - | 1e-121 | 77.49% (271) | formate/nitrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1321|M.thermoresistible__bu -----------VHFAHAR-------GYP----------TAVSDTSQRDLG
Mvan_4123|M.vanbaalenii_PYR-1 MRVCGRRAGLQAKSTHARGLHGSGCGYPGGPRRPHREDTTVSQTNQRELG
MSMEG_4318|M.smegmatis_MC2_155 ----------------------------------------MSGTSQRELG
Mflv_1015|M.gilvum_PYR-GCK ----------------------------------------MSQTSQRELG
MAV_2490|M.avium_104 ------------------MTINASAPQRQGAVAVVDTVTALPAAVAGDGG
:. : : *
TH_1321|M.thermoresistible__bu DSDSPIEDALEEAFRRMVDEGTQRLHRRWREVLVTGFFGGTEIAVGVLAY
Mvan_4123|M.vanbaalenii_PYR-1 ESDSPIEDELEYAFRRMVDEGTQRLHRSWREVLVTGFFGGTDIAVGVLAY
MSMEG_4318|M.smegmatis_MC2_155 KSNSPIEDSLEDTFNRTVDEGSQRLHRRWREILVTGFFGGTEVAMGVLAY
Mflv_1015|M.gilvum_PYR-GCK DTDSPIEDALENAFNRMLDEGTQRLHRSWHEVLVTGFFGGTEVAMGVLAY
MAV_2490|M.avium_104 TADPFEPLTVGAMVDRVSAMAVEKAAHPWAFLMRS-LVGGAMVAFGVLLA
::. : . * . :: : * :: : :.**: :*.***
TH_1321|M.thermoresistible__bu LAVLHVTHNH----LLAGLAFSIGFLALLLGRSELFTEGFLIPVTTVAAK
Mvan_4123|M.vanbaalenii_PYR-1 LAVLDATQQP----LLAALAFSVGFLALLLGRSELFTEGFLIPVTTVAAK
MSMEG_4318|M.smegmatis_MC2_155 LAVLNATGNQ----LMAGLAFSIGFLALLLGHSELFTEGFLVPVTTVAAK
Mflv_1015|M.gilvum_PYR-GCK LSVLQATGNQ----LLAGIAFSIGFLALLLGRSELFTEGFLIPVTTVAAK
MAV_2490|M.avium_104 LVVSTGVKTPGVASLLMGLAFGMSFVLILVSGMSLITADMAAGFLAVLQR
* * . *: .:**.:.*: :*:. .*:* .: . :* :
TH_1321|M.thermoresistible__bu RASVAQLMKLWGGTLAANLVGGWVLMWLIMTA---FPRLHAQTTESAAHY
Mvan_4123|M.vanbaalenii_PYR-1 RASVAQLAKLWTGTLAANLVGGWVLMWLIMTA---LPRLRAQTIESAAHY
MSMEG_4318|M.smegmatis_MC2_155 RASVAQLFKLWGGTLAANLVGGWVIMWLIMLG---FPKLHAQTVESASHF
Mflv_1015|M.gilvum_PYR-GCK RAGIGQLAKLWGGTLIANLAGGWLIMWLVMTA---YPKLHEKTTTSAEHF
MAV_2490|M.avium_104 ALSIRSYVVLVAVGLVGNIVGALVFVTVCAAAGGPYLGAFADRAATVGTQ
.: . * * .*:.*. ::: : . . :.
TH_1321|M.thermoresistible__bu ATAPLSAETMCLALLGGMVITLMTRMQHGTDAMIGKIAAAVAG--GFLLA
Mvan_4123|M.vanbaalenii_PYR-1 ATAPLSAETVALSLLGGMVITLMTRMQHGTDAMVGKIAAAVAG--AFLLA
MSMEG_4318|M.smegmatis_MC2_155 ATAPLSAETVALGLLGGMAITLMTRMQHGTDSVPGKIAAAVAG--AFLLA
Mflv_1015|M.gilvum_PYR-GCK VNAPFNAETVALALLGGMVITLMTRMQHGTDSVPGKIAAAVAG--AFLLA
MAV_2490|M.avium_104 KAGQPFWTALLLAVLCTWFLQTSMCMFFKARSDVARMALAFYGPFAFVIG
. :: *.:* : * . : : .::* *. * .*::.
TH_1321|M.thermoresistible__bu GLQMFHS---ILDSLLIFGALIAGDAP------FGYLDWLRWFGYTVVGN
Mvan_4123|M.vanbaalenii_PYR-1 GLQLFHS---ILDSLLIFGALVTGHAP------FGYLDWLGWFGYTVVGN
MSMEG_4318|M.smegmatis_MC2_155 GLQMFHS---ILDSLLIFGAIFTGEAS------FGYLDWLKWFGYTVVAN
Mflv_1015|M.gilvum_PYR-GCK GLQMFHS---ILDSLLIFGALTAGDAP------FGYLDWLSWFGYTLVLN
MAV_2490|M.avium_104 GTQHVIANVGFVGLPLLLNLFHPIAARGDIGWGFGDHGLLTNIGVTTVGN
* * . : ::. *::. : . * ** . * :* * * *
TH_1321|M.thermoresistible__bu VAGGLGLVTMLRLLRSKERLQEERRDAEAE---
Mvan_4123|M.vanbaalenii_PYR-1 IAGGLLLVTLLRLLRSKDRLREERHDADTA---
MSMEG_4318|M.smegmatis_MC2_155 VAGGLILVTLLRLLRSKERIEEEREEADAE---
Mflv_1015|M.gilvum_PYR-GCK VVGGLALVTLLRLIRSKDRLKEERRDAESPTAE
MAV_2490|M.avium_104 LIGGTVFVALPFWIIAHLQRRRLLSTGALRPDG
: ** :*:: : :: : .. .