For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDQHYLDPTAEQDRKPVLQWRVVDIVVASVLAVAAGLVFVFWNVASNPIAAPLGAVLPGLQALAGGGWLF AGVLTALVIRKPGAALYGELVAATVSALVGNQWGVLTIESGLVQGLGAELVFAAFLYRRWNLPVAVLAGA AAGVALAVNDLILWYPGSTTAFAAIYTGSAVVSGAVLAGVLSWFVVRGLAKTGALSRFASGRVPSSQSR
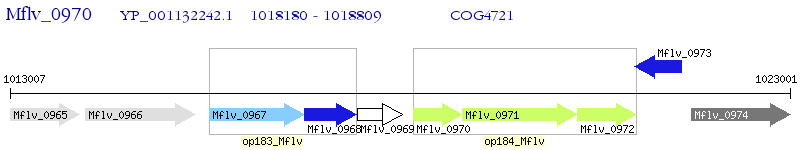
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0970 | - | - | 100% (209) | ABC transporter, permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6726 | - | 2e-89 | 82.56% (195) | ABC transporter, permease protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0970|M.gilvum_PYR-GCK ---MDQHYLDPTAEQDRKPVLQWRVVDIVVASVLAVAAGLVFVFWNVASN
MSMEG_6726|M.smegmatis_MC2_155 MTDVSRSDSGPFVPQDRPRTYRWRVVDIVVASVLAVAAGLVFVMWNIASN
:.: .* . *** . :*********************:**:***
Mflv_0970|M.gilvum_PYR-GCK PIAAPLGAVLPGLQALAGGGWLFAGVLTALVIRKPGAALYGELVAATVSA
MSMEG_6726|M.smegmatis_MC2_155 PIGAPLSAALPGLQALLGGGWLFAGVLTALVIRKPGAALYGELVAATVSA
**.***.*.******* *********************************
Mflv_0970|M.gilvum_PYR-GCK LVGNQWGVLTIESGLVQGLGAELVFAAFLYRRWNLPVAVLAGAAAGVALA
MSMEG_6726|M.smegmatis_MC2_155 LVGNQWGVLTLESGLVQGIGAELVFAVFLYRVWNLPVAILSGAVAGLALA
**********:*******:*******.**** ******:*:**.**:***
Mflv_0970|M.gilvum_PYR-GCK VNDLILWYPGSTTAFAAIYTGSAVVSGAVLAGVLSWFVVRGLAKTGALSR
MSMEG_6726|M.smegmatis_MC2_155 INDLILWYPGSAAAFTLIYTVSAVVSGALVAGALSWYAVRGLARTGALSR
:**********::**: *** *******::**.***:.*****:******
Mflv_0970|M.gilvum_PYR-GCK FASGRVPSSQSR
MSMEG_6726|M.smegmatis_MC2_155 FASGRTAAR---
*****..: