For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNAAGRLAAFGAGLAVAFTAAYVTAAAVGPDAAVTPSQHVGAEPAADHSMPAEQGVPVSPLSADPPGLSL AQDGYALSAVRAPGVPNDRATLSFTIAGPSGTPLLDYATVHDKQLHLILVRSDGQHFAHVHPVLDQATGI WSMPWMWKAAGTYRVFADFQPAQAGSAKLTLTRTVDVAGELAPSPPLTTRTSDEIAGYTVDLDGALTAGV SKQLTAKVTRDGKPVTTLQPYLGAYGHLVALREGDLAYLHVHPEGAEPVGGRTGGPAVSFAATAPTAGRY LLYLDFKVDDTVHTASFVVDAAPGGGNQPAPETPRDARHAGGR
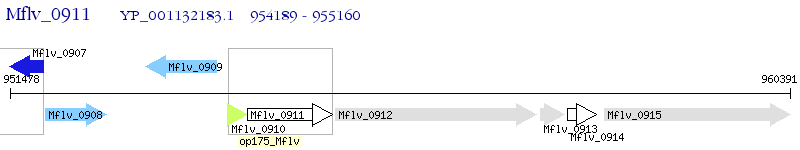
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0911 | - | - | 100% (323) | hypothetical protein Mflv_0911 |
| M. gilvum PYR-GCK | Mflv_2848 | - | e-163 | 88.20% (322) | hypothetical protein Mflv_2848 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1650c | - | 3e-11 | 25.22% (345) | hypothetical protein MAB_1650c |
| M. marinum M | MMAR_0232 | - | 7e-65 | 45.90% (305) | hypothetical protein MMAR_0232 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5015 | - | 1e-47 | 39.13% (322) | secreted protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4882 | - | 8e-62 | 51.27% (236) | hypothetical protein MUL_4882 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0232|M.marinum_M --------------------------------------------------
MUL_4882|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0911|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5015|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1650c|M.abscessus_ATCC_199 MTGLHTKCLFLVLALFTLFSGTPVALADSASAGADVQVAQSLGERELTVV
MMAR_0232|M.marinum_M --------------------------------------------------
MUL_4882|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0911|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5015|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1650c|M.abscessus_ATCC_199 ARRVTSIPGPLYVDVITHQGAPAGPLRVQLVPTGADTDEHEPAPGVPTST
MMAR_0232|M.marinum_M ---------------------------------------------MSARA
MUL_4882|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0911|M.gilvum_PYR-GCK ---------------------------------------------MNAAG
MSMEG_5015|M.smegmatis_MC2_155 ---------------------------------------------MRADA
MAB_1650c|M.abscessus_ATCC_199 EMVVLGDKPGPYSTTLDVDRPGPADLTISDGVHISRIPLVLTGQAMSPPE
MMAR_0232|M.marinum_M RLALYATG-----MAALFGAAFLVGGAVIAHPISTSWARSGHAGHQAGTQ
MUL_4882|M.ulcerans_Agy99 -------------MAALFGAAFLVGGAVIAHPISTSWARSGHAGHQAGTQ
Mflv_0911|M.gilvum_PYR-GCK RLAAFGAG-----LAVAFTAAYVTAAAVGPDAAVTPSQHVGAEPAADHSM
MSMEG_5015|M.smegmatis_MC2_155 KIAAFGAG-----LVAVFAAAFGVG-SVWADEPDAP--------------
MAB_1650c|M.abscessus_ATCC_199 RVAYVGLATTGLLLVISLVVATRVRRGVWIALPVTATVAALAVAITGAVL
:. : .* . .* :.
MMAR_0232|M.marinum_M PVR-------------GVSLSAGDLMLD---------------DVAAPPA
MUL_4882|M.ulcerans_Agy99 PVR-------------GVSLSAGDLMLD---------------DVAAPPA
Mflv_0911|M.gilvum_PYR-GCK PAEQGVPVSPLSADPPGLSLAQDGYALS---------------AVRAPGV
MSMEG_5015|M.smegmatis_MC2_155 ---------------------AAGYSLE---------------LADESVA
MAB_1650c|M.abscessus_ATCC_199 STSAPPPPEPGPQRDPTVDNAANPYRLDRPAVTDYSRPSAMLVAEQPELD
*.
MMAR_0232|M.marinum_M VGVQGTLSFRVQS-LGGVPVTRYAIENAKKLHLIVVRSDGTEFRHVHPTM
MUL_4882|M.ulcerans_Agy99 VGVQGTLSFRVQS-LGGVPVTRYAIENTKKLHLIVVRSDGTEFRHVHPTM
Mflv_0911|M.gilvum_PYR-GCK PNDRATLSFTIAG-PSGTPLLDYATVHDKQLHLILVRSDGQHFAHVHPVL
MSMEG_5015|M.smegmatis_MC2_155 PGTQVPLRFRIVD-ASGAAVTEYDENHEKDLHLIVVRDDLTGYQHLHPAL
MAB_1650c|M.abscessus_ATCC_199 PGHATDIYFSLIDTATGLPADDLVIHDSALIHLLVVAPDG-QLSHLHPIR
. : * : . * . . :**::* * *:**
MMAR_0232|M.marinum_M S-AEGRWSIPWAWNAAGTYRVFADFVPE--GGNPTVLTYALNVGGMFTPD
MUL_4882|M.ulcerans_Agy99 S-AEGRWSIPWAWNSAGTYRVFADFVPE--GGNPTVLTYALNVGGMFTPD
Mflv_0911|M.gilvum_PYR-GCK DQATGIWSMPWMWKAAGTYRVFADFQPAQAGSAKLTLTRTVDVAGELAPS
MSMEG_5015|M.smegmatis_MC2_155 D-ATGTWSVPLDVPTGGKHRVFADFVPA--GASGVVAEADLRVTGSDEPQ
MAB_1650c|M.abscessus_ATCC_199 V-APGRYSVPLSAAIAGRYAVSAEFVRRGGGVQTVRLPGALTVRGGRPAT
* * :*:* .* : * *:* * : * * .
MMAR_0232|M.marinum_M PPPS-ATNVSRTDGFTVTIDGP-FTAQSPSRLIATVTRDGQPVTTLQPYL
MUL_4882|M.ulcerans_Agy99 PPPS-ATNVSRTDGFTVTIDGP-FTAQSPSRLIATVTRDGQPVTTLQPYL
Mflv_0911|M.gilvum_PYR-GCK PPLT-TRTSDEIAGYTVDLDGA-LTAGVSKQLTAKVTRDGKPVTTLQPYL
MSMEG_5015|M.smegmatis_MC2_155 PLPA-PSNTATVDGYTVTLTGS-PVAQSASELTLSVSRDGKPAD-LQPYL
MAB_1650c|M.abscessus_ATCC_199 PAPFDGPGMRMMNGLHVHVSAAGLRAGRPTTLSATIGSE----PVLQPWL
* * * : .. * .. * .: : ***:*
MMAR_0232|M.marinum_M GAYGHLVVLR---------------EGDLAYLHAHPENGAASAGSASPAQ
MUL_4882|M.ulcerans_Agy99 GAYGHLVVLR---------------EGDLAYLHAHPENGAASAGSASPAQ
Mflv_0911|M.gilvum_PYR-GCK GAYGHLVALR---------------EGDLAYLHVHPEGAEPVGGRTGGPA
MSMEG_5015|M.smegmatis_MC2_155 GAYGHLVALR---------------ASDLHYLHVHPIG-------DSPGD
MAB_1650c|M.abscessus_ATCC_199 GMLGHLVVAGPIPRMASATDLGTAVQDSAIWAHAHSMGGHGATGMPGMEH
* ****. .. : *.*. . .
MMAR_0232|M.marinum_M IAFTTEP------PTPGRYLLYLDFQVNGRVHTA----QFVRCAAE----
MUL_4882|M.ulcerans_Agy99 IAFTTEP------PTPGRYLLYLDFQVNGRVHTA----QFVRCAAE----
Mflv_0911|M.gilvum_PYR-GCK VSFAATA------PTAGRYLLYLDFKVDDTVHTA----SFVVDAAPGGGN
MSMEG_5015|M.smegmatis_MC2_155 VAFHTEF------PSAGDYRLYFDFKHDDVVRTA----EFTVAVPEGTG-
MAB_1650c|M.abscessus_ATCC_199 SANHQSAQDDPMPPMVGVPPLNGDSPPDETVAAFGPDVPFTFTFPKPGRY
: * * * * : * : *. .
MMAR_0232|M.marinum_M ----QSSHPATHQ---------
MUL_4882|M.ulcerans_Agy99 ----QSSHPATHQ---------
Mflv_0911|M.gilvum_PYR-GCK QPAPETPRDARHAGGR------
MSMEG_5015|M.smegmatis_MC2_155 ---HQDGHQDGHQDGHSH----
MAB_1650c|M.abscessus_ATCC_199 LAWIQAQRDYRVLTIPVRLDIS
: :