For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDYALRRRSLLAEVYSGRTGVTEVCDANPYLLRAAKFHGKPSSVMCPICRKEQLTLVSWVFGDHLGAVSG SARSAEELVLLATKYDEFSVHVVEVCRTCSWNHLVKSYVLGAVPPPKGSRGPRRTQTARGTARTASE
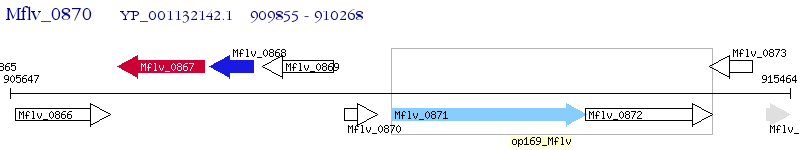
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0870 | - | - | 100% (137) | hypothetical protein Mflv_0870 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0050 | - | 9e-64 | 83.21% (137) | hypothetical protein Mb0050 |
| M. tuberculosis H37Rv | Rv0049 | - | 9e-64 | 83.21% (137) | hypothetical protein Rv0049 |
| M. leprae Br4923 | MLBr_02689 | - | 8e-58 | 77.86% (140) | hypothetical protein MLBr_02689 |
| M. abscessus ATCC 19977 | MAB_4902c | - | 7e-63 | 83.21% (137) | hypothetical protein MAB_4902c |
| M. marinum M | MMAR_0068 | - | 1e-61 | 81.02% (137) | hypothetical protein MMAR_0068 |
| M. avium 104 | MAV_0070 | - | 2e-63 | 82.14% (140) | hypothetical protein MAV_0070 |
| M. smegmatis MC2 155 | MSMEG_6901 | - | 3e-64 | 85.40% (137) | hypothetical protein MSMEG_6901 |
| M. thermoresistible (build 8) | TH_1500 | - | 3e-66 | 88.32% (137) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0067 | - | 4e-61 | 79.56% (137) | hypothetical protein MUL_0067 |
| M. vanbaalenii PYR-1 | Mvan_6037 | - | 7e-73 | 93.43% (137) | hypothetical protein Mvan_6037 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0870|M.gilvum_PYR-GCK --------------------------MDYALRRRSLLAEVYSGRTGVTEV
Mvan_6037|M.vanbaalenii_PYR-1 -------------------MRLQRQVVDYALRRRSLLAEVYSGRTGVSEV
TH_1500|M.thermoresistible__bu -------------------------VVDYALRRRSLLADVYSGRTGVYEV
MAB_4902c|M.abscessus_ATCC_199 -------------------MRLQRQVVDYALKRRSLLAEVYSGRTGVAEV
MSMEG_6901|M.smegmatis_MC2_155 MSCDAVAPRCPTGRRTLVNVRLQRQVVDYALRRRSLLAEVYSGRTGVSEV
Mb0050|M.bovis_AF2122/97 --------------------------MDYTLRRRSLLAEVYSGRTGVSEV
Rv0049|M.tuberculosis_H37Rv --------------------------MDYTLRRRSLLAEVYSGRTGVSEV
MMAR_0068|M.marinum_M -------------------MRLQRQVVDYALRRRSLLAEVYSGRTGVSEV
MUL_0067|M.ulcerans_Agy99 -------------------MRLQRQVLDYALRRRSLLAEVYSGRTGVSEV
MAV_0070|M.avium_104 -----------------MGVRLQRQVVDYALRRRSLLAEVYSGRTGVSEV
MLBr_02689|M.leprae_Br4923 -------------------------MVNYSLRRRFLLAEVYSGRTGVSEV
::*:*:** ***:******** **
Mflv_0870|M.gilvum_PYR-GCK CDANPYLLRAAKFHGKPSSVMCPICRKEQLTLVSWVFGDHLGAVSGSARS
Mvan_6037|M.vanbaalenii_PYR-1 CDANPYLLRAAKFHGKPSTVMCPICRKEQLTLVSWVFGDHLGAVSGSART
TH_1500|M.thermoresistible__bu CDANPYLLRAAKFHGKPSSVMCPICRKEQLTLVSWVFGDHLGAVSGSART
MAB_4902c|M.abscessus_ATCC_199 CDANPYLLRAAKFHGKPSAVVCPICRKEQLTLVSWVFGEHLGAVSGSART
MSMEG_6901|M.smegmatis_MC2_155 CDANPYLLRAAKFHGKQSSVMCPICRKEQLTLVSWVFGEHLGPVSGSART
Mb0050|M.bovis_AF2122/97 CDANPYLLRAAKFHGKPSRVICPICRKEQLTLVSWVFGEHLGAVSGSART
Rv0049|M.tuberculosis_H37Rv CDANPYLLRAAKFHGKPSRVICPICRKEQLTLVSWVFGEHLGAVSGSART
MMAR_0068|M.marinum_M CDANPYLLRAAKFHGKQSQVICPICRKEQLTLVSWVFGDHLGPVSGSART
MUL_0067|M.ulcerans_Agy99 CDANPYLLRAAKFHGKQSQVICPICRKEQLTLVSWVFGDHLGPVSGSART
MAV_0070|M.avium_104 CDANPYLLRAAKFHGKPSQIMCPICRKEQLTLVSWVFGEHLGPVSGSART
MLBr_02689|M.leprae_Br4923 CDANPYLLRAAKFHGKPSQVMCPICRKEQLTLVSWVFGNQLGAISGSART
**************** * ::*****************::**.:*****:
Mflv_0870|M.gilvum_PYR-GCK AEELVLLATKYDEFSVHVVEVCRTCSWNHLVKSYVLGAV---PPPKGSRG
Mvan_6037|M.vanbaalenii_PYR-1 AEELVLLATRYDEFSVHVVEVCRTCSWNHLVKSYVLGAI---PPPKGSRG
TH_1500|M.thermoresistible__bu AEELVVLASRYDEFSVHVVEVCRTCSWNHLVKSYVLGKT---RPPKGTR-
MAB_4902c|M.abscessus_ATCC_199 AEELVLLATRYSEFAVHVVEVCRTCSWNHLVKSYVMGAL---PAPKGTT-
MSMEG_6901|M.smegmatis_MC2_155 AEELVLLASRYDEFAVHVVEVCRTCSWNHLVKSYVLGAP---RPPEARR-
Mb0050|M.bovis_AF2122/97 AEELILLATRFSEFAVHVVEVCRTCSWNHLVKSYVLGAARPARPPRGS--
Rv0049|M.tuberculosis_H37Rv AEELILLATRFSEFAVHVVEVCRTCSWNHLVKSYVLGAARPARPPRGS--
MMAR_0068|M.marinum_M AEELIMLTTRFSEFAVHVVEVCRTCSWNHLVKSYVLGAARPARPPRGT--
MUL_0067|M.ulcerans_Agy99 AEELIMLTTRFSEFAVHVVEVCRTCSWNHLVKSYVLGAARPARPPRGT--
MAV_0070|M.avium_104 AEELVMLATRFEEFAVHVVEVCRTCSWNHLVKSYVLGAARPAPPPRGTR-
MLBr_02689|M.leprae_Br4923 AEELVLLATRYEEFAVYVVEVCRTCSWNYLVRSYVLGAARSAPPPRGT--
****::*::::.**:*:***********:**:***:* .*..
Mflv_0870|M.gilvum_PYR-GCK PRRTQTARGTARTASE
Mvan_6037|M.vanbaalenii_PYR-1 SRSTRTARGAARTASE
TH_1500|M.thermoresistible__bu TRGTRTARDGARTASE
MAB_4902c|M.abscessus_ATCC_199 P-RQRTTR--ARTASE
MSMEG_6901|M.smegmatis_MC2_155 PKRTPSTRKSARTASE
Mb0050|M.bovis_AF2122/97 -GGTRTARNGARTASE
Rv0049|M.tuberculosis_H37Rv -GGTRTARNGARTASE
MMAR_0068|M.marinum_M -GGKRTARNGARTASE
MUL_0067|M.ulcerans_Agy99 -GGKRTARNGGRTASE
MAV_0070|M.avium_104 -TTTRTARNGARTATE
MLBr_02689|M.leprae_Br4923 -PVTRTACNGARMAIE
:: .* * *