For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVLGIVVVALVIAVGLVGIVVPILPGGILVIGAIGVWAFVVNTTVSWIVFGVAAALFAASQVIKYTWPVK RMRAADVRTSVLVVGGIAGVVGFFVIPVIGLLIGFVLGVFAAEMSIRGDATRAWASTWHALKGVALSVGV ELTGALLATVAWVVGVVLTL*
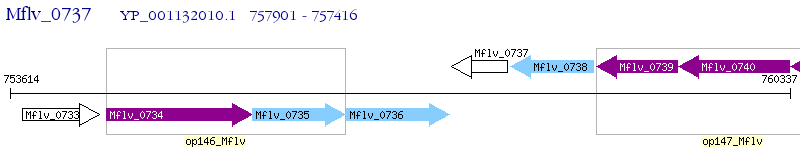
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0737 | - | - | 100% (161) | hypothetical protein Mflv_0737 |
| M. gilvum PYR-GCK | Mflv_0983 | - | 4e-05 | 26.90% (145) | hypothetical protein Mflv_0983 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4592c | - | 2e-43 | 50.31% (159) | hypothetical protein MAB_4592c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0099 | - | 8e-62 | 71.25% (160) | hypothetical protein MSMEG_0099 |
| M. thermoresistible (build 8) | TH_4082 | - | 4e-55 | 60.62% (160) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0112 | - | 8e-75 | 86.25% (160) | hypothetical protein Mvan_0112 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0737|M.gilvum_PYR-GCK MSVLGIVVVALVIAVGLVGIVVPILPGGILVIGAIGVWAFVVNTTVSWIV
Mvan_0112|M.vanbaalenii_PYR-1 MSTGGIVLVALAIAIGLVGIIVPILPGGILVIGAIGVWAFVDNTAVAWVV
MSMEG_0099|M.smegmatis_MC2_155 MSTVGIVLVAFAIAVGLIGIVVPILPGGLLVFGAIAVWAIVERTAAAWVV
TH_4082|M.thermoresistible__bu MSTGGVILVALAVAVGIVGIVVPILPGTLLIFAAIGVWAFVEASPAAWVT
MAB_4592c|M.abscessus_ATCC_199 MSVGGLLLVALGIAIGMVGIVVPMVPGTGLVVLSIGAWALVESTTKAWVV
**. *:::**: :*:*::**:**::** *:. :*..**:* :. :*:.
Mflv_0737|M.gilvum_PYR-GCK FGVAAALFAASQVIKYTWPVKRMRAADVRTSVLVVGGIAGVVGFFVIPVI
Mvan_0112|M.vanbaalenii_PYR-1 LGVAAALFIASQIIKYTWPVKRMRRADVRTSVLVVGGVAGVVGFFVIPVI
MSMEG_0099|M.smegmatis_MC2_155 LGVAAAVFVAAEVIKYLWPVRRMRAAEVGTWSLVAGGVLGVIGFFVIPVI
TH_4082|M.thermoresistible__bu FGVVTALLGAGQLIKYLWPMRRMRRADVSTSTLVVGAVVGIIGFFVIPVL
MAB_4592c|M.abscessus_ATCC_199 LGIVVGLAATTTVVKYVWPAARMREAGVPTLSLVLGGIAAIVGFFVIPVV
:*:...: : ::** ** *** * * * ** *.: .::*******:
Mflv_0737|M.gilvum_PYR-GCK GLLIGFVLGVFAAEMSIRGDATRAWASTWHALKGVALSVGVELTGALLAT
Mvan_0112|M.vanbaalenii_PYR-1 GLLIGFVLGVYAAELGIRRDAARAWASTWHALKGVALSVGVELTGALLAT
MSMEG_0099|M.smegmatis_MC2_155 GLVIGFVAGVYLAELAARHEHRRAWVSTVHALKGVALSVGVELTGALLAT
TH_4082|M.thermoresistible__bu GLVLGFVGGVYLAELGRRGDARAAWSATVHAIKGVALSVGLELAAALLAT
MAB_4592c|M.abscessus_ATCC_199 GLVVGFVLGIYLAEFIRRSSQRQAWGATVTAVKAVATSIGIEMVGAMVSA
**::*** *:: **: * . ** :* *:*.** *:*:*:..*::::
Mflv_0737|M.gilvum_PYR-GCK VAWVVGVVLTL
Mvan_0112|M.vanbaalenii_PYR-1 VAWVVGVFLTA
MSMEG_0099|M.smegmatis_MC2_155 IAWVVGVVVTN
TH_4082|M.thermoresistible__bu AVWVAGLFVT-
MAB_4592c|M.abscessus_ATCC_199 GIWLGAVLV--
*: .:.: