For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQSPSPAPGWYPDPLNPGKQIYWDGASWGEPVGVPDNSSNAKKTGVAIGVCVLAVVGLVMSMQSVSLMTG SGPVWTGVAVVGVATAIAFFMGAATWVRVLACVLLALSLVNGFYIESQMSEKRDELTRVFDN*
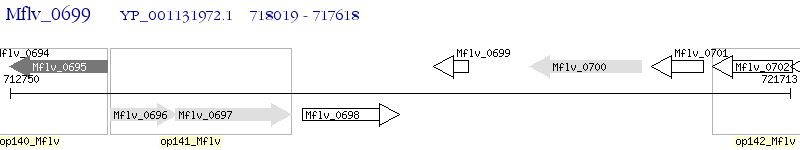
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0699 | - | - | 100% (133) | hypothetical protein Mflv_0699 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3061 | - | 7e-06 | 38.98% (59) | hypothetical protein MAV_3061 |
| M. smegmatis MC2 155 | MSMEG_1312 | - | 8e-05 | 28.87% (97) | hypothetical protein MSMEG_1312 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0999 | - | 3e-44 | 62.12% (132) | hypothetical protein MUL_0999 |
| M. vanbaalenii PYR-1 | Mvan_0765 | - | 4e-07 | 36.92% (65) | hypothetical protein Mvan_0765 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0699|M.gilvum_PYR-GCK -----MSQSPSPAPGWYPDPLN-PGKQIYWDGASWG--------EPVGVP
MUL_0999|M.ulcerans_Agy99 -----MT-APLPAPGWYPDPSS-PEKQIYWDGTAWS--------GPP-AS
MSMEG_1312|M.smegmatis_MC2_155 -----MTAPNTPAAGWYPDPSG-QPGQRYHDGRRWTQHFVPT--PPPAPA
MAV_3061|M.avium_104 MPSEPAPPPPKSAPGWYPDPAVGHGLKRYFDGTSWTGEWAFF--TEQRKN
Mvan_0765|M.vanbaalenii_PYR-1 --------MTQPTAGWYPDPSD-PSRQRYFDGKTWTENYAPFPAPPPGIG
.:.****** : * ** *
Mflv_0699|M.gilvum_PYR-GCK DNSSNAKKTGVAIG---------VCVLAVVGLVMSMQSVSLMTG------
MUL_0999|M.ulcerans_Agy99 GAADKHKKTAVAIG---------VCVLVLVGLVMSMQSVSLMSG------
MSMEG_1312|M.smegmatis_MC2_155 APPPAPVAVAVATGGGPSHGLHAVLTLLTCGMWLPIWILVAIFGGGGTSA
MAV_3061|M.avium_104 GTSGKAALALAALG--------VVVLLLIFAKSWAFDPEKAARATATSAA
Mvan_0765|M.vanbaalenii_PYR-1 QPAKPGMSRGLKIGLGVG---AAVLALVAIGNIGGNEKSAETRSSSTSPS
. * * * . .
Mflv_0699|M.gilvum_PYR-GCK -----SGPVWTG-------------VAVVGVATAIAFFM-----------
MUL_0999|M.ulcerans_Agy99 -----SGPVWTG-------------VGVVAAGTAVAFFL-----------
MSMEG_1312|M.smegmatis_MC2_155 VAVSGAGVVRTANRR---------PAMVVGAVIGGLLLL-----------
MAV_3061|M.avium_104 TGSTEPAPPTPAGPKKPDGVTFTTAAAPDGDVVDARFAIRDNYSEEMIRD
Mvan_0765|M.vanbaalenii_PYR-1 SSFSESSTSTVTSASDDD-----APAPAGSTVRDGKFEFQVLGVERG---
.. . . : :
Mflv_0699|M.gilvum_PYR-GCK ----GAA---TWVRVLACVLLALSLVNGFYIESQMSEKRDELTRVFDN--
MUL_0999|M.ulcerans_Agy99 ----RAATWATWVRVVAALVLAVSLANALYIENEIAKKREQLTHIFDR--
MSMEG_1312|M.smegmatis_MC2_155 ----GLVGQHPWLLVVFGVLGVVAGFGMWLLKSAQRREEQQRAEQFQRDM
MAV_3061|M.avium_104 GARSDTIQILKYAHATYPNASAVNVQGTFPMTDPYGNTSTQVAIDLTYSR
Mvan_0765|M.vanbaalenii_PYR-1 ---ATSIDDAFGPEVAKGEFFTVRLRVTNIGDDARSFSATNQKLIINGNE
. .: . : :
Mflv_0699|M.gilvum_PYR-GCK --------------------------------------------------
MUL_0999|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1312|M.smegmatis_MC2_155 LAQ-RAEYEDQLYQQGDPRGVHGRYMPPPTP-------------------
MAV_3061|M.avium_104 ATLNKINFDGVRANTIWEIRDSGTVLPAFQP-------------------
Mvan_0765|M.vanbaalenii_PYR-1 YEATSIMDDGWMEDINPGLGIEASATFDIPPGAVPSAIECHDSMFSGGAL
Mflv_0699|M.gilvum_PYR-GCK ---
MUL_0999|M.ulcerans_Agy99 ---
MSMEG_1312|M.smegmatis_MC2_155 ---
MAV_3061|M.avium_104 ---
Mvan_0765|M.vanbaalenii_PYR-1 LAL