For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDSRQRRITALTEAAKVKSQNKTRDAEQAIRRLIKRGDPITFQAVQREAGVSHAFLYNHPELRTRIEHLR GTRRKATADQPVDTDSTLVITLTRQIAELKKQHRQQLQALRAALERAHGENLDLRRELARRGIQPSPNVV SIATTS*
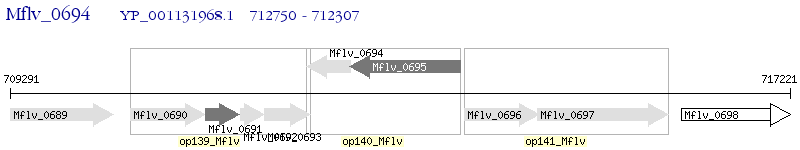
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0694 | - | - | 100% (147) | Tn554-related, transposase C |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0444 | - | 3e-12 | 36.97% (119) | putative transposase/integrase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4157 | - | 7e-08 | 30.40% (125) | putative transposase |
| M. thermoresistible (build 8) | TH_4584 | - | 3e-06 | 29.23% (130) | PUTATIVE putative hypothetical protein Mjls_1428 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5933 | - | 1e-69 | 89.80% (147) | Tn554-related, transposase C |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0694|M.gilvum_PYR-GCK MTDSRQRRITALTEAAKVKSQNKTRDAEQAIRRLIKRGDPITFQAVQREA
Mvan_5933|M.vanbaalenii_PYR-1 MTDSRERRIKALTEAAKAKSQNKTRDAEQAIRRLVKRGGPITFQAVQREA
MAB_0444|M.abscessus_ATCC_1997 --MSTPRSTEGLRRHTTQLRASTARKIKQAIREMKKKGLPINPNSVSRYS
TH_4584|M.thermoresistible__bu --VTPPRSTEGLLDSTKRRKAATRVKVRKALREMKKKGRTININAVANYA
MSMEG_4157|M.smegmatis_MC2_155 ---MRADNSIHIVTAAKQRHELTRAKAIAALHELDRAGTKISFEAVAEHA
: :. . . *::.: : * *. ::* . :
Mflv_0694|M.gilvum_PYR-GCK GVSHAFLYNHPELRTRIEHLRGTRRKATADQP---VDTDSTLVITLTRQI
Mvan_5933|M.vanbaalenii_PYR-1 GVSHAFLYNHPELRKRIEHLRGARRKAPADQS---VDTDSMLVITLTRQI
MAB_0444|M.abscessus_ATCC_1997 GVSRKTIYNHTDLLTQIRAAANKPVIRPADQGEGPPTTESGIIAALRNQI
TH_4584|M.thermoresistible__bu KVARKTIYNHPDLRDEIRAAATAPRPRPTTPPGDEANGETSITAALRDQL
MSMEG_4157|M.smegmatis_MC2_155 GVSRSWLYTQPDLKDEINRIRALRRPQHDQAP---PARQRATEDSLRQRL
*:: :*.:.:* .*. : :* ::
Mflv_0694|M.gilvum_PYR-GCK AELKKQHRQQLQALR-------AALERAHGENLDLRRELARRGIQPSPNV
Mvan_5933|M.vanbaalenii_PYR-1 AELKKQHRQQLQALR-------DALERAHGENLDLRRELARRGTDPTPNV
MAB_0444|M.abscessus_ATCC_1997 TALKTGHRADIAELKATIKGLNEDLAAAHGEIYTLAEQLRHRGKSSIDKG
TH_4584|M.thermoresistible__bu RSQKTRYETDIATLKSQVKQLQQELATAHGELH----RLRNAG-GRPQRG
MSMEG_4157|M.smegmatis_MC2_155 D-VALRHNRELAEQN---QRLRHQLAHALGQARDDTRKRPARHRDSITID
:. :: . * * *: .
Mflv_0694|M.gilvum_PYR-GCK VSIATTS
Mvan_5933|M.vanbaalenii_PYR-1 TSIATTS
MAB_0444|M.abscessus_ATCC_1997 SGTASPG
TH_4584|M.thermoresistible__bu TS-----
MSMEG_4157|M.smegmatis_MC2_155 PC-----