For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAVTLEILQPGPLALVEDLGRPGLSHIGVTRSGAADRRAHTLANRLVANPDDRATVEVTLGGFTARVHD GNGDGVAIAVTGADANPSVNGVPFGTNSIHHAHDGEVISLAPPRSGLRSYLAVRGGIDVRPVLGSRSYDV MSAIGPGPLRAGDVLPVGPHTDDFPELDQAPVAAIDDDFLELKVVPGPRDDWFVDPDVLIRTNWQVTNRS DRVGMRLVGMPLEYRSPDRQLPSEGATRGAIQVPPNGFPVILGPDHPVTGGYPVIGVVTDDDIDKIAQAR PGQTVRLHWSRPRRPWES
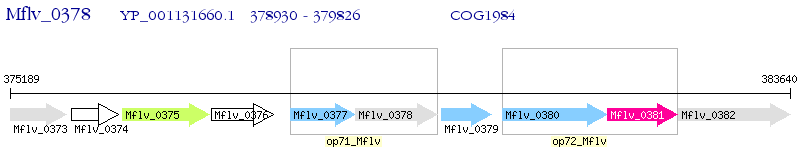
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0378 | - | - | 100% (298) | urea amidolyase related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0269c | - | 1e-131 | 76.82% (289) | hypothetical protein Mb0269c |
| M. tuberculosis H37Rv | Rv0263c | - | 1e-131 | 76.82% (289) | hypothetical protein Rv0263c |
| M. leprae Br4923 | MLBr_02550 | - | 1e-122 | 73.61% (288) | hypothetical protein MLBr_02550 |
| M. abscessus ATCC 19977 | MAB_4394 | - | 1e-119 | 70.24% (289) | hypothetical protein MAB_4394 |
| M. marinum M | MMAR_0523 | - | 1e-129 | 75.26% (291) | urea amidolyase-related protein |
| M. avium 104 | MAV_4897 | - | 1e-129 | 76.74% (288) | hypothetical protein MAV_4897 |
| M. smegmatis MC2 155 | MSMEG_0435 | - | 1e-145 | 82.99% (294) | allophanate hydrolase subunit 2 |
| M. thermoresistible (build 8) | TH_0925 | - | 1e-141 | 79.93% (294) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1186 | - | 1e-127 | 74.23% (291) | urea amidolyase-related protein |
| M. vanbaalenii PYR-1 | Mvan_0303 | - | 1e-156 | 89.23% (297) | urea amidolyase related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0378|M.gilvum_PYR-GCK MSAVT---LEILQPGPLALVEDLGRPGLSHIGVTRSGAADRRAHTLANRL
Mvan_0303|M.vanbaalenii_PYR-1 MNGVT---LEILQPGPLALVEDLGRPGMSHIGVTRSGAADRRSHTLANRL
MSMEG_0435|M.smegmatis_MC2_155 -MGTT---LEVLRTGPLALVEDLGRPGLAHMGVTRSGAADRRSHTLANRL
TH_0925|M.thermoresistible__bu -MTVT---LEVLRTGPLALIEDLGRPGLAHLGVSPSGAADRRSHMLANRL
MMAR_0523|M.marinum_M -MIT----LEILRSGPLAIVEDLGRAGLGHLGVGRSGAADRRSHTLANRL
MUL_1186|M.ulcerans_Agy99 -MIT----LEILRSGPLAIVEDLGRAGLGHLGVGRSGAADRRSHTLANRL
Mb0269c|M.bovis_AF2122/97 -MTT----LEILRSGPLALVEDLGRAGLAHLGVGRSGAADRRSHTLANRL
Rv0263c|M.tuberculosis_H37Rv -MTT----LEILRSGPLALVEDLGRAGLAHLGVGRSGAADRRSHTLANRL
MAV_4897|M.avium_104 -MPF----LEILRTGPLAVVEDLGRPGLAHLGVSRSGAADRRAHKLANRL
MLBr_02550|M.leprae_Br4923 -MTNPPIKLEILRSGPFAVVQDLGRAGLAHFGVGRSGAADRRAHKLANRL
MAB_4394|M.abscessus_ATCC_1997 -MRPT---LEVMRTGPLALVQDLGRVGKAAMGVGRSGAADRSSHSLANRL
**:::.**:*:::**** * . :** ****** :* *****
Mflv_0378|M.gilvum_PYR-GCK VANPDDRATVEVTLGGFTARVHDGNGDGVAIAVTGADANPSVNGVPFGTN
Mvan_0303|M.vanbaalenii_PYR-1 VANPDDWATVEVTMGGFTARVHGGDGEGVAIAVTGADADPAVNGIPFGTN
MSMEG_0435|M.smegmatis_MC2_155 VANPGESATIEVTFGGFSARVCGGD---VAIAVTGADTDPAVNGIPFGTN
TH_0925|M.thermoresistible__bu VANPTDRATIEVMFGGFSARVRGGD---VAIAVTGADANPAVNGIPFGTN
MMAR_0523|M.marinum_M VANPDDWATVEVTLGGFSARVRGGD---IDIAVTGADTDPTVNGTLFGTN
MUL_1186|M.ulcerans_Agy99 VANPDDWATVEVTLGGFSARVRGGD---IDIAVTGADTDPTVNGALFGTN
Mb0269c|M.bovis_AF2122/97 VANPDDWATVEVTFGGFSARVRGGD---VDIAVTGADTDPTVNGIMVGTN
Rv0263c|M.tuberculosis_H37Rv VANPDDWATVEVTFGGFSARVRGGD---VDIAVTGADTDPTVNGIMVGTN
MAV_4897|M.avium_104 VANPDDRATVEVTFGGFAARVRGGD---VDIAVTGADADPTVDGIEFGSN
MLBr_02550|M.leprae_Br4923 VANPDDRATVEVTFGGFSARVCGGN---VYVAVTGADTNPTANGIVFGTN
MAB_4394|M.abscessus_ATCC_1997 VANPDNLATIEVTLGGMSFRVIGGD---VVVAVTGADTDPSINGIPFGTN
**** : **:** :**:: ** .*: : :******::*: :* .*:*
Mflv_0378|M.gilvum_PYR-GCK SIHHAHDGEVISLAPPRSGLRSYLAVRGGIDVRPVLGSRSYDVMSAIGPG
Mvan_0303|M.vanbaalenii_PYR-1 SIHYARDGEVISLGSPRSGLRSYLAVRGGIDVEPVLGSRSYDVMSMIGPQ
MSMEG_0435|M.smegmatis_MC2_155 SIHHVHDGQVISLGAPHSGLRSYLAVRGGIDVTPVLGSRSYDVMSAIGPS
TH_0925|M.thermoresistible__bu SIQYVHDGEVISLGSPRTGLRSYLAVRGGIDVEPTLGSRSHDVLAGIGPR
MMAR_0523|M.marinum_M SIHHVRDGSVISLGTPNAGLRTYLAVRGGISVTPVLGSRSYDVMSAIGPA
MUL_1186|M.ulcerans_Agy99 SIHHVRDGSVISLGTPNAGLRTYLSVRGGISVTPVLGSRSYDVMSAIGPA
Mb0269c|M.bovis_AF2122/97 SIHHVRDGQVISLGTPRAGLRTYLAVRGGVCVEPVLGSRSYDVMSAIGPS
Rv0263c|M.tuberculosis_H37Rv SIHHVRDGQVISLGTPRAGLRTYLAVRGGVCVEPVLGSRSYDVMSAIGPS
MAV_4897|M.avium_104 SIQHVRDGQVIALGTPRAGLRSYLAVRGGICVDPVLGSRSYDVMSAIGPA
MLBr_02550|M.leprae_Br4923 SIQHIRDGQVISLGAPHVGLRTYLAVRGGICVTPVLGSRSYDVLSAIGPA
MAB_4394|M.abscessus_ATCC_1997 SIAQVQDGDVVSLGAPRSGLRSYVAVRGGVSVAPVMGSRSFDIMSGIGPK
** :**.*::*..*. ***:*::****: * *.:****.*::: ***
Mflv_0378|M.gilvum_PYR-GCK PLRAGDVLPVGPHTDDFPELDQAPVAAIDDDFLELKVVPGPRDDWFVDPD
Mvan_0303|M.vanbaalenii_PYR-1 PLRRGDVLTVGAHTDDFPELEQAPVAAIVDDVLELKVVPGPRDDWFVDPD
MSMEG_0435|M.smegmatis_MC2_155 PLRPGDVLPVGEHTDEFPELDQAPVAAIAEDVVELQVVPGPRDDWFVDPD
TH_0925|M.thermoresistible__bu PLKAGDVLPVGTQSDEFPELDQAPVAAIDEDLVEVAVMPGPRDDWFVDPD
MMAR_0523|M.marinum_M PLRTGDVLPIGDRSQDYPELDQAPVAAITDRVVELRAIPGPRDDWFVDPE
MUL_1186|M.ulcerans_Agy99 PLRTGDVLPIGDRSQDYPELDQAPVAAITDRVVELRAIPGPRDDWFVDPE
Mb0269c|M.bovis_AF2122/97 PLRAGDVLPVGEHTDDYPELDQAPVAAIEEHLVELRVVPGPRDDWLVDPD
Rv0263c|M.tuberculosis_H37Rv PLRAGDVLPVGEHTDDYPELDQAPVAAIEEHLVELRVVPGPRDDWLVDPD
MAV_4897|M.avium_104 PLRAGDRLPIGEHTQDYPELDQAPVAAITGHLMELQVVPGPRDDWLVNPD
MLBr_02550|M.leprae_Br4923 PLRAGDELPVGDHTDHYPALDHAPVAAITGNPVELLVIPGPHDDWFIDPD
MAB_4394|M.abscessus_ATCC_1997 PLRAGDRLPIGPRSPSFPEVEQAPVATISDDVVELKVVPGPRDDWFTDPD
**: ** *.:* :: :* :::****:* :*: .:***:***: :*:
Mflv_0378|M.gilvum_PYR-GCK VLIRTNWQVTNRSDRVGMRLVGMPLEYRSPDRQLPSEGATRGAIQVPPNG
Mvan_0303|M.vanbaalenii_PYR-1 VLVRTNWQVTNRSDRVGMRLVGMPLDYRWPDRQLPSEGATRGAIQVPPNG
MSMEG_0435|M.smegmatis_MC2_155 ILVRTNWLVTNRSDRVGMRLVGMPLEYRNPDRQLPSEGATRGAIQVPPNG
TH_0925|M.thermoresistible__bu VLVRTNWLATNRSDRVGMRLVGMPLEYREPDRQLPSEGATRGAVQVPPNG
MMAR_0523|M.marinum_M ALVHTDWVVSGQSDRVGTRLVGRPLTHRNPDRQLPSEGTTRGAIQVPPNG
MUL_1186|M.ulcerans_Agy99 ALVHTDWVVSGQSDRVGNRLVGRPLTHRNPDRQLPSEGTTRGAIQLSPNG
Mb0269c|M.bovis_AF2122/97 ALVHTIWMASNRSDRVGMRLQGRPLQHRWPDRQLPGEGVTRGAIQVPPNG
Rv0263c|M.tuberculosis_H37Rv ALVHTIWMASNRSDRVGMRLQGRPLQHRWPDRQLPGEGVTRGAIQVPPNG
MAV_4897|M.avium_104 ALVHTIWMASDRSDRVGMRLEGRPLQHRYPDRQLPSEGTTRGAIQVPPNG
MLBr_02550|M.leprae_Br4923 VLVHTDWVASDRSDRVGMRLLGSPLLYRYPDRQLPSEGTTCGAIQLPPNG
MAB_4394|M.abscessus_ATCC_1997 ALIHGNWIVSDRSDRVGTRLIGKPLELRDPARQLPSEGVVRGAIQVPPGG
*:: * .:.:***** ** * ** * * ****.**.. **:*:.*.*
Mflv_0378|M.gilvum_PYR-GCK FPVILGPDHPVTGGYPVIGVVTDDDIDKIAQARPGQTVRLHWSRPRRPWE
Mvan_0303|M.vanbaalenii_PYR-1 FPVILGPDHPVTGGYPVIGVVVDDDIDKVAQARPGQTVRLHWSRPRRPFE
MSMEG_0435|M.smegmatis_MC2_155 FPVILGPDHPVTGGYPVIGVVTEEDIDKLGQVRPGQTVRLHWAYPRRPFE
TH_0925|M.thermoresistible__bu FPVILGPDHPVTGGYPVIGVVVDEDVDKIAQVRPGQTVRLHWARPRRPFD
MMAR_0523|M.marinum_M LPVILGPDHPITGGYPVVGVVIDDDIDKVAQVRPGQHVWLHWARPRR---
MUL_1186|M.ulcerans_Agy99 LPVILGPDHPITGGYPVVGVVIDDDIDKVAQVRPGQHVWLHWARPRR---
Mb0269c|M.bovis_AF2122/97 LPVILGPDHPITGSYPVVGVITDEDIDKVAQIRPGQYVRLHWARPRSRLP
Rv0263c|M.tuberculosis_H37Rv LPVILGPDHPITGSYPVVGVITDEDIDKVAQIRPGQYVRLHWARPRSRLP
MAV_4897|M.avium_104 LPVILGPDHPVTGGYPVVGVVTDEDVDKIAQVRPGQYVRLHWARPRALAG
MLBr_02550|M.leprae_Br4923 LPVILGPDHPVTGGYPVAGVVIDEDIDKLAQLRPGQQVRLHWVRTQSSPL
MAB_4394|M.abscessus_ATCC_1997 QPVILGPDHPVTGGYPVIGVITDHDVDLAAQVRPGQTVRFSWSRPRMN--
*********:**.*** **: :.*:* .* **** * : * .:
Mflv_0378|M.gilvum_PYR-GCK S----------------
Mvan_0303|M.vanbaalenii_PYR-1 DDGRAVNWALTPPDDRW
MSMEG_0435|M.smegmatis_MC2_155 T----------------
TH_0925|M.thermoresistible__bu HQADPVMAAPLPQPW--
MMAR_0523|M.marinum_M -----------------
MUL_1186|M.ulcerans_Agy99 -----------------
Mb0269c|M.bovis_AF2122/97 GQGVTQAW---------
Rv0263c|M.tuberculosis_H37Rv GQGVTQAW---------
MAV_4897|M.avium_104 AHAGAASWPFA------
MLBr_02550|M.leprae_Br4923 ERLRSASQN--------
MAB_4394|M.abscessus_ATCC_1997 -----------------