For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KVTCNDEAVEVDDGTTVTQLLGLLGIPEKGIAVAVDWAVIPRSHWDTALTDGAKVEVVTAVQGG*
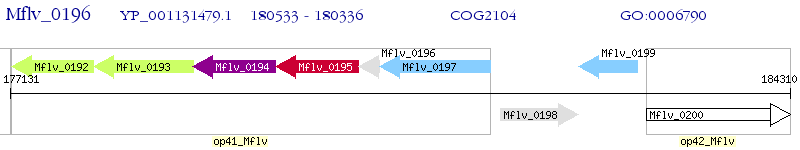
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0196 | - | - | 100% (65) | sulfur carrier protein ThiS |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0424 | thiS | 1e-12 | 48.53% (68) | sulfur carrier protein ThiS |
| M. tuberculosis H37Rv | Rv0416 | thiS | 1e-12 | 48.53% (68) | sulfur carrier protein ThiS |
| M. leprae Br4923 | MLBr_00298 | - | 8e-13 | 47.30% (74) | sulfur carrier protein ThiS |
| M. abscessus ATCC 19977 | MAB_4217c | - | 1e-15 | 55.38% (65) | putative thiamine biosynthesis protein ThiS |
| M. marinum M | MMAR_0719 | thiS | 4e-16 | 58.46% (65) | thiamine biosynthesis protein ThiS |
| M. avium 104 | MAV_4745 | thiS | 2e-15 | 58.46% (65) | sulfur carrier protein ThiS |
| M. smegmatis MC2 155 | MSMEG_0792 | thiS | 4e-19 | 63.49% (63) | sulfur carrier protein ThiS |
| M. thermoresistible (build 8) | TH_2313 | thiS | 2e-23 | 72.31% (65) | thiamine biosynthesis protein ThiS |
| M. ulcerans Agy99 | MUL_2804 | thiS | 3e-16 | 58.46% (65) | thiamine biosynthesis protein ThiS |
| M. vanbaalenii PYR-1 | Mvan_0709 | - | 4e-26 | 84.62% (65) | sulfur carrier protein ThiS |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0196|M.gilvum_PYR-GCK -------MKVTCNDEAVEVDDGTTVTQLLGLLGIPEKGIAVAVDWAVIPR
Mvan_0709|M.vanbaalenii_PYR-1 -------MKVTVNDELVEVDDATTVAALLDRLGFPEKGIAVAVDWAVIPR
MSMEG_0792|M.smegmatis_MC2_155 ------MITITVNGESVEVDDTITIERLLETRGFPEKGIAVALDWSVLHR
TH_2313|M.thermoresistible__bu MKADMKTMRVTLNDETIEVDEQTTVATLLEQRGFPEKGIAVAVDWSVLPR
MMAR_0719|M.marinum_M -------MIVVVNERRVEVEEQTTVAALLQSLGFPDRGIAVALDLAVLPR
MUL_2804|M.ulcerans_Agy99 -------MIVVVNERRVEVEEQTTVAALLQSLGFPDRGIAVALDLAVLPR
Mb0424|M.bovis_AF2122/97 -------MIVVVNEQQVEVDEQTTIAALLDSLGFGDRGIAVALNFSVLPR
Rv0416|M.tuberculosis_H37Rv -------MIVVVNEQQVEVDEQTTIAALLDSLGFGDRGIAVALNFSVLPR
MLBr_00298|M.leprae_Br4923 -------MIVVVNERPVEVNEQTTVAALLESLGFPASGIAVAVEFSVLPR
MAV_4745|M.avium_104 -------MIVVVNEQKVDVDAHTTVAALLDELGFGGRGVAVAVDDAVLPR
MAB_4217c|M.abscessus_ATCC_199 -------MNIQVNGDAREVTEGANIWQLLEQLGFPESGIAVAINHTVVPK
: : * :* .: ** *: *:***:: :*: :
Mflv_0196|M.gilvum_PYR-GCK SHWDTALTDG---------AKVEVVTAVQGG
Mvan_0709|M.vanbaalenii_PYR-1 SQWDTALTDG---------AKVEVVAAVQGG
MSMEG_0792|M.smegmatis_MC2_155 SEWDQTLSDG---------ARIEVVTAVQGG
TH_2313|M.thermoresistible__bu SRWDTTLTDG---------ARIEVVTAVQGG
MMAR_0719|M.marinum_M SDWATELSEG---------DRLEVVTAVQGG
MUL_2804|M.ulcerans_Agy99 SDWATELSEG---------DRLEVVTAVQGG
Mb0424|M.bovis_AF2122/97 SDWATKICEL-----R-KPVRLEVVTAVQGG
Rv0416|M.tuberculosis_H37Rv SDWATKICEL-----R-KPVRLEVVTAVQGG
MLBr_00298|M.leprae_Br4923 SYWATKISELPAVTGRSEPIRLEVVTAVQGG
MAV_4745|M.avium_104 SRWTTELSEG---------ARLEVVTAVQGG
MAB_4217c|M.abscessus_ATCC_199 SDWDTVVSVG---------ATVEVVTAVQGG
* * : :***:*****