For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GEGRTGDVTRRLWSVQLQGLVGYVLFALQVAVLVASVYSFVHAAMQRPDAYTAADKLTKPVWLVILGVGV LLALVLGITGVAIAAVAAGVYIVDVRPKILEVQGKSR*
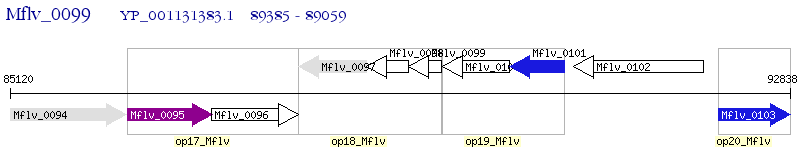
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0099 | - | - | 100% (108) | hypothetical protein Mflv_0099 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0486 | - | 2e-29 | 70.11% (87) | transmembrane protein |
| M. tuberculosis H37Rv | Rv0476 | - | 2e-29 | 70.11% (87) | transmembrane protein |
| M. leprae Br4923 | MLBr_02453 | - | 1e-31 | 68.48% (92) | hypothetical protein MLBr_02453 |
| M. abscessus ATCC 19977 | MAB_4082c | - | 8e-19 | 50.57% (87) | hypothetical protein MAB_4082c |
| M. marinum M | MMAR_0801 | - | 9e-33 | 72.83% (92) | transmembrane protein |
| M. avium 104 | MAV_4674 | - | 8e-30 | 70.11% (87) | hypothetical protein MAV_4674 |
| M. smegmatis MC2 155 | MSMEG_0920 | - | 3e-27 | 62.77% (94) | hypothetical protein MSMEG_0920 |
| M. thermoresistible (build 8) | TH_0870 | - | 4e-31 | 74.16% (89) | POSSIBLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4544 | - | 1e-31 | 71.74% (92) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_0809 | - | 2e-44 | 93.55% (93) | hypothetical protein Mvan_0809 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0099|M.gilvum_PYR-GCK MGEGRTGDVTRRLWSVQLQGLVGYVLFALQVAVLVASVYSFVHAAMQRPD
Mvan_0809|M.vanbaalenii_PYR-1 ---------------MQLQGLVGYVLFALQIAVLVTSAYAFVHAAMQRPD
TH_0870|M.thermoresistible__bu ---------------VSSH-IVGTVLFLLTMAVLAVTVYAFVHAAMQRPD
MMAR_0801|M.marinum_M -----------------MSQLVGTVLLVLQIAVLVTAVYAFVHAALQRPD
MUL_4544|M.ulcerans_Agy99 -----------------MSQLVGTVLLVLQIAVLVTAVYAIVHAAMQRPD
MLBr_02453|M.leprae_Br4923 -----------------MSHLVGTVMLVLQLAVLVTAVYAFVHAALQRPD
MAV_4674|M.avium_104 -------------------------MLVLQIAVLVTAIYAFVHAALQRPD
Mb0486|M.bovis_AF2122/97 -------------------------MLVLLVAVLVTAVYAFVHAALQRPD
Rv0476|M.tuberculosis_H37Rv -------------------------MLVLLVAVLVTAVYAFVHAALQRPD
MSMEG_0920|M.smegmatis_MC2_155 ---------------MILADLAGVIILVLVALVFFVGVYALVHAAMQRPD
MAB_4082c|M.abscessus_ATCC_199 ------------------MAFSFHVFALISLITLIAAIVALVHAAIQPAD
: : .: . ::****:* .*
Mflv_0099|M.gilvum_PYR-GCK AYTAADKLTKPVWLVILGVGVLLAL----VLGITGVAIAAVAAGVYIVDV
Mvan_0809|M.vanbaalenii_PYR-1 AYTAADKLTKPVWLVILGVGVLLAL----VLGITGVAIAAVAAGVYLVDV
TH_0870|M.thermoresistible__bu AFTAADKLTKPVWLVILGVSALLTFLP--VLSILGPVIAAVASGVYLVDV
MMAR_0801|M.marinum_M AYTAADKLTKPVWLVILGAAVALTSILGFVFGVLGMAIAACAAGVYLVDV
MUL_4544|M.ulcerans_Agy99 AYTAADKLTKPVWLVILGAAVALTSILGFVFGVLGMAIAACAAGVYLVDV
MLBr_02453|M.leprae_Br4923 AYTAAEKLTKPVWLVILGAAVSLTSILGFVFGVLGIVIAACAAGVYLVDV
MAV_4674|M.avium_104 AYTAADKLTKPVWLLILGGAVALTSVLGYVFGVLGMAIAACAAGVYLVDV
Mb0486|M.bovis_AF2122/97 AYTAADKLTKPVWLVILGAAVALASILYPVLGVLGMAMSACASGVYLVDV
Rv0476|M.tuberculosis_H37Rv AYTAADKLTKPVWLVILGAAVALASILYPVLGVLGMAMSACASGVYLVDV
MSMEG_0920|M.smegmatis_MC2_155 AYTATGKLTKPVWLIILGVGLLLLLLLRDAF---GAAISACATGVYLVDV
MAB_4082c|M.abscessus_ATCC_199 AFVAAEKQTKTTWVVILAGAAVLTLIPG--LELLTAGIAAVAAGVYFVDV
*:.*: * **..*::**. . * : ::* *:***:***
Mflv_0099|M.gilvum_PYR-GCK RPKILEVQGKSR
Mvan_0809|M.vanbaalenii_PYR-1 RPKILEIQGKSR
TH_0870|M.thermoresistible__bu RPKILEIQGKSH
MMAR_0801|M.marinum_M RPKLLEIQGKSR
MUL_4544|M.ulcerans_Agy99 RPKLLEIQDKSR
MLBr_02453|M.leprae_Br4923 RPKLLDIQGKSR
MAV_4674|M.avium_104 RPKLLEIQGKSR
Mb0486|M.bovis_AF2122/97 RPKLLEIQGKSR
Rv0476|M.tuberculosis_H37Rv RPKLLEIQGKSR
MSMEG_0920|M.smegmatis_MC2_155 RPKLLEIQGKSR
MAB_4082c|M.abscessus_ATCC_199 RPRVLEVQGKSR
**::*::*.**: