For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQVTSVGHAGFLIQTQAGSILCDPWVNPAYFASWFPFPDNSGLDWGALGECDYLYVSHLHKDHFDAENLR AHVNKDAVVLLPDFPVPDLRNELQKLGFHRFFETTDSVKHRLRGPNGDLDVMIIALRAPADGPIGDSALV VADGETTAFNMNDARPVDLDVLASEFGHIDVHMLQYSGAIWYPMVYDMPARAKDAFGAQKRQRQMDRARQ YIAQVGATWVVPSAGPPCFLAPELRHLNDDGSDPANIFPDQMVFLDQMRAHGQDGGLLMIPGSTADFTGT TLNSLRHPLPAEQVEAIFTTDKAAYIADYADRMAPVLAAQKAGWAAAAGEPLLQPLRTLFEPIMLQSNEI CDGIGYPVELAIGPETIVLDFPKRAVREPIPDERFRYGFAIAPELVRTVLRDNEPDWVNTIFLSTRFRAW RVGGYNEYLYTFFKCLTDERIAYADGWFAEAHDDSSSITLNGWEIQRRCPHLKADLSKFGVVEGNTLTCN LHGWQWRLDDGRCLTARGHQLRSSRP
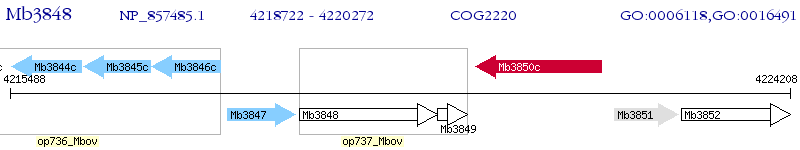
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3848 | - | - | 100% (516) | hypothetical protein Mb3848 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1152 | - | 0.0 | 84.01% (519) | Rieske (2Fe-2S) domain-containing protein |
| M. tuberculosis H37Rv | Rv3818 | - | 0.0 | 100.00% (516) | hypothetical protein Rv3818 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0156c | - | 0.0 | 80.97% (515) | Rieske family iron-sulfur cluster-binding protein |
| M. marinum M | MMAR_5381 | - | 0.0 | 89.71% (515) | hypothetical protein MMAR_5381 |
| M. avium 104 | MAV_0200 | - | 0.0 | 86.80% (515) | Rieske 2Fe-2S family protein |
| M. smegmatis MC2 155 | MSMEG_6410 | - | 0.0 | 82.47% (519) | Rieske 2Fe-2S family protein |
| M. thermoresistible (build 8) | TH_0894 | - | 0.0 | 82.17% (516) | HYPOTHETICAL PROTEIN Rv3818 |
| M. ulcerans Agy99 | MUL_5001 | - | 0.0 | 88.54% (515) | hypothetical protein MUL_5001 |
| M. vanbaalenii PYR-1 | Mvan_5657 | - | 0.0 | 83.43% (519) | Rieske (2Fe-2S) domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0156c|M.abscessus_ATCC_199 MRVTSVGHAGFFIETAAGSILCDPWVNPAYFASWFPFPDNSTLDWDRFGS
TH_0894|M.thermoresistible__bu VQVTSIGHAGFRLDTTAGSILCDPWVNPAYFGSWFPFPDNSTLDWDALGD
MSMEG_6410|M.smegmatis_MC2_155 MQVTSVGHAGFLIESRAGSILCDPWVNPAYFASWFPFPDNSQLDWDALGD
Mflv_1152|M.gilvum_PYR-GCK MQVTSVGHAGFRIDTKAGSILCDPWVNPAYFASWFPFPDNTGLDWPALGA
Mvan_5657|M.vanbaalenii_PYR-1 MQVTSVGHAGFRIDTKAGSILCDPWLNPAYFASWFPFPDNSDLDWAALGD
Mb3848|M.bovis_AF2122/97 MQVTSVGHAGFLIQTQAGSILCDPWVNPAYFASWFPFPDNSGLDWGALGE
Rv3818|M.tuberculosis_H37Rv MQVTSVGHAGFLIQTQAGSILCDPWVNPAYFASWFPFPDNSGLDWGALGE
MMAR_5381|M.marinum_M MQVTSVGHAGFLIQTQSGSILCDPWVNPAYFASWFPFPDNSALDWDALGD
MUL_5001|M.ulcerans_Agy99 MQVTSVGHAGFLIQTQSGSILCDPWVNPAYFASWFPFPDNSALDWDAIGD
MAV_0200|M.avium_104 MQVTSVGHAGFLIRTQAGSILCDPWVNPAYFASWFPFPDNSTLDWDQLGD
::***:***** : : :********:*****.********: *** :*
MAB_0156c|M.abscessus_ATCC_199 CDYLYVSHLHKDHYDPKNLTDHVNKDAVVLLPDYPVPDLRRELEGLGFHT
TH_0894|M.thermoresistible__bu VDYLYVSHLHKDHFDPELLRAHVRKDAVVLLPDFPVPDLRRELEKLGFHR
MSMEG_6410|M.smegmatis_MC2_155 VDYLYVSHLHKDHFDPEHLRRYVNKDAVVLLPDYPVPDLRRELEKLGFHN
Mflv_1152|M.gilvum_PYR-GCK VDYLYVSHLHKDHFDPKNLAEHVNKDAVVLLPDFPVPDLRRELEKLGFHT
Mvan_5657|M.vanbaalenii_PYR-1 VDYLYVSHLHKDHFDPQNLRDHVNKDAVVLLPDFPVPDLRRELEKLGFHR
Mb3848|M.bovis_AF2122/97 CDYLYVSHLHKDHFDAENLRAHVNKDAVVLLPDFPVPDLRNELQKLGFHR
Rv3818|M.tuberculosis_H37Rv CDYLYVSHLHKDHFDAENLRAHVNKDAVVLLPDFPVPDLRNELQKLGFHR
MMAR_5381|M.marinum_M CDYLYVSHLHKDHFDAENLRSHVNKDAVVLLPDFPVPDLRNELEKLGFHR
MUL_5001|M.ulcerans_Agy99 CDYLYVSHLHKDHFDAENLRSHVNKDAVVLLPDFPVPDLRNELEKLGFHR
MAV_0200|M.avium_104 CDYLYVSHLHKDHFDAKNLAEHVNKDAVVLLPEFPVPDLRNALQELGFHR
************:*.: * :*.********::******. *: ****
MAB_0156c|M.abscessus_ATCC_199 FVETEDSVKHRVSGPKGDLDIMIIALRAPADGPIGDSGLVVSDGTTTAFN
TH_0894|M.thermoresistible__bu FFETTDSVKHTVSGPKGELEVMIVALRAPADGPIGDSGLVVSDGTTTVFN
MSMEG_6410|M.smegmatis_MC2_155 FFETTDSVKHTVSGPKGDLDVMIIALRAPADGPIGDSGLVVSDRVTTVFN
Mflv_1152|M.gilvum_PYR-GCK FFETTDSVKHRVSGPKGDLDVMIIALRAPADGPIGDSGLVVSDGVTTAFN
Mvan_5657|M.vanbaalenii_PYR-1 FFETADSVKHRVSGPKGELDVMIVALRAPADGPIGDSGLVVDDGETTVFN
Mb3848|M.bovis_AF2122/97 FFETTDSVKHRLRGPNGDLDVMIIALRAPADGPIGDSALVVADGETTAFN
Rv3818|M.tuberculosis_H37Rv FFETTDSVKHRLRGPNGDLDVMIIALRAPADGPIGDSALVVADGETTAFN
MMAR_5381|M.marinum_M FFETTDSVKHRLSGPRGDLDIMIIALRAPADGPIGDSALVVSDGETTAFN
MUL_5001|M.ulcerans_Agy99 FFETTDSVKHRLSGPRSDLDIMIIALRAPADGPIGDSALVVSDGETTAFN
MAV_0200|M.avium_104 FFETADSVKHRVGG----LDVMIIALRAPADGPIGDSALVVSDGSTTLFN
*.** ***** : * *::**:*************.*** * ** **
MAB_0156c|M.abscessus_ATCC_199 MNDARPVDLDVLAEEFGHIDVHMLQYSGAIWYPMVYDMPARAKEAFGTQK
TH_0894|M.thermoresistible__bu MNDARPVDLDVLAVEFGHIDVHMLQFSGAIWYPMVYDMPKRAKAAFGTQK
MSMEG_6410|M.smegmatis_MC2_155 MNDARPVDLDVLHTDFGQIDVHMLQYSGAIWYPMVYDMPARAKEAFGIQK
Mflv_1152|M.gilvum_PYR-GCK MNDARPVDLDMVHADFGQIDVHMLQYSGAIWYPMVYDMPARAKQAFGTQK
Mvan_5657|M.vanbaalenii_PYR-1 MNDARPVDLDMLHTDFGQIDVHMLQYSGAIWYPMVYDMPARAKEAFGIQK
Mb3848|M.bovis_AF2122/97 MNDARPVDLDVLASEFGHIDVHMLQYSGAIWYPMVYDMPARAKDAFGAQK
Rv3818|M.tuberculosis_H37Rv MNDARPVDLDVLASEFGHIDVHMLQYSGAIWYPMVYDMPARAKDAFGAQK
MMAR_5381|M.marinum_M MNDARPIELDMLASEFGHIDVHLLQYSGAIWYPMVYDMPARAKEAFGTQK
MUL_5001|M.ulcerans_Agy99 MNDARPIELDMLASEFGHVDVHLLQYSGAIWYPMVYDMPARAKEAFGTQK
MAV_0200|M.avium_104 MNDARPVELDVLASEFGHIDVHLLQYSGAIWYPMVYDMPARAKESFGIQK
******::**:: :**::***:**:************* *** :** **
MAB_0156c|M.abscessus_ATCC_199 RQRQMDRCRQYIAQVGATWVIPSAGPPCFLDTGLRDLNDDHDDPANIFPD
TH_0894|M.thermoresistible__bu RQRQMDRCRQYIDQVGATWVIPSAGPPCFLDPDLRHLNDDSGDPANIFPD
MSMEG_6410|M.smegmatis_MC2_155 RQRQMDRCRQYIAQVGATWVVPSAGPPCFLDPELRDLNDDHGDPANIFPD
Mflv_1152|M.gilvum_PYR-GCK RQRQMDRCRQYIAQVGATWVIPSAGPPCFLDPELRDLNDDHGDPANIFPD
Mvan_5657|M.vanbaalenii_PYR-1 RQRQMDRCRQYIAQVGATWVIPSAGPPCFLDPELRGLNDDHGDPANIFPD
Mb3848|M.bovis_AF2122/97 RQRQMDRARQYIAQVGATWVVPSAGPPCFLAPELRHLNDDGSDPANIFPD
Rv3818|M.tuberculosis_H37Rv RQRQMDRARQYIAQVGATWVVPSAGPPCFLAPELRHLNDDGSDPANIFPD
MMAR_5381|M.marinum_M RQRGMDRARQYVAQVDATWVVPSAGPPCFLDPELRELNDDRSDPANIFPD
MUL_5001|M.ulcerans_Agy99 RQRGMDRARQYVAQVDATWVVPSAGPPCFLDPELRELNDDRSDPANIFPD
MAV_0200|M.avium_104 RQRQMDRARQYLAQVGATWVVPSAGPPCFLDPELRHLNDDHGDPANIFPD
*** ***.***: **.****:********* . ** **** .********
MAB_0156c|M.abscessus_ATCC_199 QLVFLDQMRQHGHDKGLLMIPGTVADFTASELNSLEHPVSTEEAEDIFGA
TH_0894|M.thermoresistible__bu QMVFLDQLRSHGHTGGLLMIPGTKAEFTGPQLDSLVHPLPEHEVTAIFTT
MSMEG_6410|M.smegmatis_MC2_155 QMVFLEQLRIHGHDGGLLMIPGSTADFTGSTLNSLTHPVDDPES--IFTT
Mflv_1152|M.gilvum_PYR-GCK QVVFLDQMRSHGHDGGLLMIPGSTADFTGSQLLSLKHPIPDDDVMAIFTT
Mvan_5657|M.vanbaalenii_PYR-1 QVVFLDQMRTHGHDGGLLMIPGSAADFAGSQLRSLTHPIPDDDVQAIFTT
Mb3848|M.bovis_AF2122/97 QMVFLDQMRAHGQDGGLLMIPGSTADFTGTTLNSLRHPLPAEQVEAIFTT
Rv3818|M.tuberculosis_H37Rv QMVFLDQMRAHGQDGGLLMIPGSTADFTGTTLNSLRHPLPAEQVEAIFTT
MMAR_5381|M.marinum_M QMVFLDQMRIHGHDRGLLMIPGSIADFTGSTLNSLSHPLPTEDVEAIFTT
MUL_5001|M.ulcerans_Agy99 QMVFLDQMRIHGHDRGLLVIPGSIGDFTGSTLNSLSHPLPTEDVEAIFTT
MAV_0200|M.avium_104 QMVFLEQMRAHGQGGGLLMIPGSTADFSGSTLNSLHHPLPTAEVEAIFTT
*:***:*:* **: ***:***: .:*:.. * ** **: : ** :
MAB_0156c|M.abscessus_ATCC_199 GKAAYIEAYAQRMAPVLAAEKSSWAPAEGEPLLAPLRAKFEPIMAQTDQI
TH_0894|M.thermoresistible__bu GKADYIDAYAQRMAPVLAAEKARWAPAAGPSLLEPLRAKFEPIMAQSDQV
MSMEG_6410|M.smegmatis_MC2_155 GKAAYIEDYAQRMAPVLAAEKARWAPSAGEPMLEALRALFEPIMTQTDQI
Mflv_1152|M.gilvum_PYR-GCK GKADYIADFAERMAPVLAAEKASWAPAEGEPLLEPLRELFEPIMLQSDQI
Mvan_5657|M.vanbaalenii_PYR-1 GKADYIADYAERMAPVLAAERASWAPAAGKPLLEPLRELFEPIMLASDQI
Mb3848|M.bovis_AF2122/97 DKAAYIADYADRMAPVLAAQKAGWAAAAGEPLLQPLRTLFEPIMLQSNEI
Rv3818|M.tuberculosis_H37Rv DKAAYIADYADRMAPVLAAQKAGWAAAAGEPLLQPLRTLFEPIMLQSNEI
MMAR_5381|M.marinum_M GKSAYIADYADRMAPVIAAERASWAPAEGEPLLEPLRALFEPIMLQSNEI
MUL_5001|M.ulcerans_Agy99 GKSAYIADYADRMAPVIAAERASWAPAEGEPLLEPLRALFEPIMLQSNEI
MAV_0200|M.avium_104 GKADYIAAYAERMAPVIAAERAGWAPATGEPLLEPLRALFEPIMSQSDEI
.*: ** :*:*****:**::: **.: * .:* .** ***** ::::
MAB_0156c|M.abscessus_ATCC_199 CDGIGYPVELRLG----SETVVLDFPKRIVREPIPNEKYRYGFEIAPELV
TH_0894|M.thermoresistible__bu CDGIGYPVELRLHGPEHRETVVLDFPKRIVREPIPNEKFRYGFAIAPELV
MSMEG_6410|M.smegmatis_MC2_155 CDGIGYPVELRLTGRDHNETVVLDFPKRVVREPIPDEKFRYGFEIPAALV
Mflv_1152|M.gilvum_PYR-GCK CDGIGYPVELRLGDGDRQEVVVLDFPKRAVREAIPDEKFRYGFAIAPELV
Mvan_5657|M.vanbaalenii_PYR-1 CDGIGYPVELRLHSRGHEETIVLDFPKRAVREAIPDEKFRYGFAIAPELV
Mb3848|M.bovis_AF2122/97 CDGIGYPVELAIG----PETIVLDFPKRAVREPIPDERFRYGFAIAPELV
Rv3818|M.tuberculosis_H37Rv CDGIGYPVELAIG----PETIVLDFPKRAVREPIPDERFRYGFAIAPELV
MMAR_5381|M.marinum_M CDGIGYPVELVIG----AETVVLDFPKRAVREPIPNEKFRYGFAIAPELV
MUL_5001|M.ulcerans_Agy99 CDGIGYPVELVIG----AETVVLDFPKRAVREPIPNEKFRYGFAIAPELV
MAV_0200|M.avium_104 CDGIGYPVELVLG----PERVILDFPKRTVREPIPDEKVRYGFTIAPELV
********** : * ::****** ***.**:*: **** *.. **
MAB_0156c|M.abscessus_ATCC_199 RTVLRDDEPDWVNTIFLSTRFKAWRVGGYNEYLYTFFKCLTDERIAYADG
TH_0894|M.thermoresistible__bu RTVLRDDEPDWVNTIFLSTRFTAWRVGGYNEYLYTFFKCLTDERIAYADG
MSMEG_6410|M.smegmatis_MC2_155 RTVLRDEEPDWVNTIFLSTRFRAWRVGGYNEYLYTFFKCLTDERIAYADG
Mflv_1152|M.gilvum_PYR-GCK RTVLRDGEPDWVNTIFLSTRFRAWRVGGYNEYLYTFFKCLTDERIAYADG
Mvan_5657|M.vanbaalenii_PYR-1 RTVVRDREPDWVNTIFLSTRFTAWRVGGYNEYLYTFFKCLTDERIAYADG
Mb3848|M.bovis_AF2122/97 RTVLRDNEPDWVNTIFLSTRFRAWRVGGYNEYLYTFFKCLTDERIAYADG
Rv3818|M.tuberculosis_H37Rv RTVLRDNEPDWVNTIFLSTRFRAWRVGGYNEYLYTFFKCLTDERIAYADG
MMAR_5381|M.marinum_M RTVLRDQEPDWVNTIFLSTRFQAWRVGGYNEYLYTFFKCLTDERIAYADG
MUL_5001|M.ulcerans_Agy99 RTVLRDQEPDWVNTIFLSTRFQAWRVGGYNEYLYTFFKCLTDDRIAYADG
MAV_0200|M.avium_104 RTVLRDREPDWVNTIFLSTRFKAWRVGGYNEYLYTFFKCLTDERIAYADG
***:** ************** ********************:*******
MAB_0156c|M.abscessus_ATCC_199 WFAEAHDDSASITLDGWEIQRRCPHLKADLSKFGVVEGKNLTCNLHGWEW
TH_0894|M.thermoresistible__bu WFAEAHDDTASITLDGWEIQRRCPHLKADLSKFGVVEGTTLTCNLHGWQW
MSMEG_6410|M.smegmatis_MC2_155 WFAEAHDDSSSITLDGFQIQRRCPHLKADLSKFGVVEGNTLTCNLHGWQW
Mflv_1152|M.gilvum_PYR-GCK WFAEAHDDTASITLEGWEIQRRCPHLKADLSKFGVIEGTTLTCNLHGWQW
Mvan_5657|M.vanbaalenii_PYR-1 WFAEAHDDCASITLDGWEIQRRCPHLKADLSKFGVVEGNTLTCNLHGWQW
Mb3848|M.bovis_AF2122/97 WFAEAHDDSSSITLNGWEIQRRCPHLKADLSKFGVVEGNTLTCNLHGWQW
Rv3818|M.tuberculosis_H37Rv WFAEAHDDSSSITLNGWEIQRRCPHLKADLSKFGVVEGNTLTCNLHGWQW
MMAR_5381|M.marinum_M WFAETHDDSASITLDGWEIQRRCPHLKADLSKFGVVEGDTLTCNLHGWQW
MUL_5001|M.ulcerans_Agy99 WFAETHDDSASITLDGWEIQRRCPHLKADLSKFGVVEGDTLTCNLHGWQW
MAV_0200|M.avium_104 WFAETHDNSASITLDGWQIQRRCPHLKADLSKFGVVEGNTLTCNLHGWQW
****:**: :****:*::*****************:** .********:*
MAB_0156c|M.abscessus_ATCC_199 NLENGRCLTSKGHELRSRKLG-
TH_0894|M.thermoresistible__bu DLRTGRCLTTKGHELRCASLPS
MSMEG_6410|M.smegmatis_MC2_155 NLENGRCLTTKGHELRCQKL--
Mflv_1152|M.gilvum_PYR-GCK NLTNGRCLTTKGHELRSTKLG-
Mvan_5657|M.vanbaalenii_PYR-1 NLTNGRCLTTNGHELRSAKL--
Mb3848|M.bovis_AF2122/97 RLDDGRCLTARGHQLRSSRP--
Rv3818|M.tuberculosis_H37Rv RLDDGRCLTARGHQLRSSRP--
MMAR_5381|M.marinum_M RLEDGRCLTTRGHQLRSSRK--
MUL_5001|M.ulcerans_Agy99 RLEDGRCLTTRGHQLRSSRK--
MAV_0200|M.avium_104 RLDDGRCLTAKGHQLRSSRA--
* *****:.**:**.