For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSLTRRPPKSPPQRPPRISGVVRLRLDIAYDGTDFAGWAAQVGQRTVAGDLDAALTTIFRTPVRLRAAGR TDAGVHASGQVAHVDVPADALPNAYPRAGHVGDPEFLPLLRRLGRFLPADVRILDITRAPAGFDARFSAL RRHYVYRLSTAPYGVEPQQARYITAWPRELDLDAMTAASRDLMGLHDFAAFCRHREGATTIRDLQRLDWS RAGTLVTAHVTADAFCWSMVRSLVGALLAVGEHRRATTWCRELLTATGRSSDFAVAPAHGLTLIQVDYPP DDQLASRNLVTRDVRSG
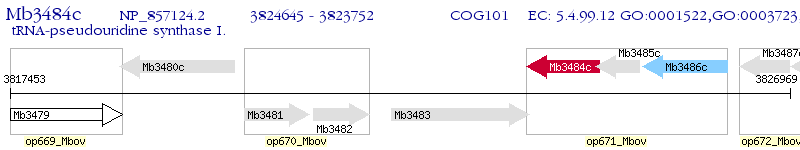
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3484c | truA | - | 100% (297) | tRNA pseudouridine synthase A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4968 | truA | 1e-121 | 71.33% (293) | tRNA pseudouridine synthase A |
| M. tuberculosis H37Rv | Rv3455c | truA | 1e-173 | 100.00% (297) | tRNA pseudouridine synthase A |
| M. leprae Br4923 | MLBr_01955 | truA | 1e-113 | 77.24% (246) | putative pseudouridylate synthase |
| M. abscessus ATCC 19977 | MAB_3768c | - | 1e-105 | 69.20% (276) | tRNA pseudouridine synthase A |
| M. marinum M | MMAR_1092 | truA | 1e-114 | 83.76% (234) | pseudouridine synthase a, TruA |
| M. avium 104 | MAV_4395 | truA | 1e-122 | 83.20% (250) | tRNA pseudouridine synthase A |
| M. smegmatis MC2 155 | MSMEG_1527 | truA | 1e-116 | 72.92% (277) | tRNA pseudouridine synthase A |
| M. thermoresistible (build 8) | TH_0034 | truA | 1e-120 | 76.09% (276) | PROBABLE TRNA PSEUDOURIDINE SYNTHASE A TRUA (PSEUDOURIDYLATE |
| M. ulcerans Agy99 | MUL_0850 | truA | 1e-124 | 82.81% (256) | tRNA pseudouridine synthase A |
| M. vanbaalenii PYR-1 | Mvan_1438 | truA | 1e-121 | 76.00% (275) | tRNA pseudouridine synthase A |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4968|M.gilvum_PYR-GCK MKPSSQPVNEPATDSGGGFV--------RLRLDIAYDGTDFAGWATQAGQ
Mvan_1438|M.vanbaalenii_PYR-1 -------MNEPATDSGGGLVFISSRPGVRLRLDIAYDGTDFAGWATQAGQ
MSMEG_1527|M.smegmatis_MC2_155 ---------MPAIESGGGHV--------RLRLDIAYDGTDFAGWAVQAGQ
TH_0034|M.thermoresistible__bu ---------MPAIDSGGGHV--------RLRLDIAYDGTDFAGWATQAGQ
Mb3484c|M.bovis_AF2122/97 ------MSLTRRPPKSPPQRPPRISGVVRLRLDIAYDGTDFAGWAAQVGQ
Rv3455c|M.tuberculosis_H37Rv ------MSLTRRPPKSPPQRPPRISGVVRLRLDIAYDGTDFAGWAAQVGQ
MMAR_1092|M.marinum_M --------------------------------------------------
MUL_0850|M.ulcerans_Agy99 ---------------------------------------------MQTGQ
MLBr_01955|M.leprae_Br4923 --------------------------------------------------
MAV_4395|M.avium_104 --------------------------------------------------
MAB_3768c|M.abscessus_ATCC_199 ---MVTSHDQPAIPDGGLVR---------LRLDIAYDGTDFAGWATQVNQ
Mflv_4968|M.gilvum_PYR-GCK RTVAGVIDDALSTVFRTPVLTRAAGRTDSGVHATGQVAHADIPTDALPHA
Mvan_1438|M.vanbaalenii_PYR-1 RTVAGAIDDALSTVFRTPVVTRAAGRTDSGVHATGQVAHVDVPADALPYA
MSMEG_1527|M.smegmatis_MC2_155 RTVAGVIDEALTTVFRTPVVTRVAGRTDTGVHATGQVAHVDVPAEALPHA
TH_0034|M.thermoresistible__bu RTVAGVLEEALSTVFRTALQLRGAGRTDTGVHATGQVAHVDVPADALVHA
Mb3484c|M.bovis_AF2122/97 RTVAGDLDAALTTIFRTPVRLRAAGRTDAGVHASGQVAHVDVPADALPNA
Rv3455c|M.tuberculosis_H37Rv RTVAGDLDAALTTIFRTPVRLRAAGRTDAGVHASGQVAHVDVPADALPNA
MMAR_1092|M.marinum_M ------------------MRLRAAGRTDAGVHATGQVAHADVPVDALPNA
MUL_0850|M.ulcerans_Agy99 RTVAGVLDEALTTVLRTPVRLRAAGRTDAGVHATGQVAHADVPVDALPNA
MLBr_01955|M.leprae_Br4923 -----MLDEAFTTVFRTPVRLRAAGRTDTGVHATGQVAHVDIPGTALSKA
MAV_4395|M.avium_104 --MAGVLDEALTTVFRTPVRLRAAGRTDAGVHATGQVAHLDVPGTALPNA
MAB_3768c|M.abscessus_ATCC_199 RTVAGVLGEAFTTIVGDSVQLYAAGRTDAGVHATGQVAHLDVPVACLPGL
: *****:****:***** *:* .*
Mflv_4968|M.gilvum_PYR-GCK YPRTVRAGEPEFGPLVRRLGRMLTEDVRVRGIVRAAPGFDARFSALRRHY
Mvan_1438|M.vanbaalenii_PYR-1 YPRTTRPGEAEFGPLVRRLGRMLPEDVRVREITRAAPGFDARFSALRRHY
MSMEG_1527|M.smegmatis_MC2_155 YPRHTRPTDAEFTPLVRRLAGFLPADVRVRDIVRAPAGFDARFSALRRHY
TH_0034|M.thermoresistible__bu YPRTGRPAGHEFLPLVRRLARFLPTDVRVLDIRRAAPGFDARFSALRRHY
Mb3484c|M.bovis_AF2122/97 YPRAGHVGDPEFLPLLRRLGRFLPADVRILDITRAPAGFDARFSALRRHY
Rv3455c|M.tuberculosis_H37Rv YPRAGHVGDPEFLPLLRRLGRFLPADVRILDITRAPAGFDARFSALRRHY
MMAR_1092|M.marinum_M YSRSARAGDPEFLPLVRRLARFLPADLRVLDIARAPTAFDARFSALRRHY
MUL_0850|M.ulcerans_Agy99 YSRSARAGDPEFLPLVRRLARFLPADLRVLDIARAPTAFDARFSALRRHY
MLBr_01955|M.leprae_Br4923 NPRSPHTGDPEFLPLLRRLSRLLPVDVRVLDIACAPVGFDARFSALRRHY
MAV_4395|M.avium_104 YPRAPHVGEAEFGPLLRRLGRFLPTDVRVVDITRAPAGFDARFSALRRHY
MAB_3768c|M.abscessus_ATCC_199 RGRYRPAGAPEFDRLVRRLSRFLPSDVRVLDISRAPAHFDARFSALRRHY
* ** *:***. :*. *:*: * *. ************
Mflv_4968|M.gilvum_PYR-GCK VYRLTTAPYGVEPAEARFVTAWPRPLDVDAMAGAAQNVLGLHDFAAFCRH
Mvan_1438|M.vanbaalenii_PYR-1 LYRLSTAPYGVEPAEARFVTPWPRPLDVAAMAAAARELVGLHDFAAFCRH
MSMEG_1527|M.smegmatis_MC2_155 VYRLTTAPYGPEPHETRFVTPWTRPLDVDAMNAAAQELLGLNDFAAFCRY
TH_0034|M.thermoresistible__bu IYRLSVAPYGVMPQLARYVTVWPRPLDVDAMAAASAQLVGLHDFAAFCRH
Mb3484c|M.bovis_AF2122/97 VYRLSTAPYGVEPQQARYITAWPRELDLDAMTAASRDLMGLHDFAAFCRH
Rv3455c|M.tuberculosis_H37Rv VYRLSTAPYGVEPQQARYITAWPRELDLDAMTAASRDLMGLHDFAAFCRH
MMAR_1092|M.marinum_M VYRLSTAPYGVEPQQARYVTVWPRELDIDAMTVASRELLGLHNFAAFCRR
MUL_0850|M.ulcerans_Agy99 VYRLSTAPYGVEPQQARYVTVWPRELDIDAMTVASQELLGLHNFAAFCRR
MLBr_01955|M.leprae_Br4923 RYRLSTAPYGVQPNQARYVTAWPRELDLDSMSAASRDLLGLRDFAVFCRH
MAV_4395|M.avium_104 VYRLSTAPWGVLPQQARYVTAWPRPLDVEAMAAASRDLLGLHDFAAFCRH
MAB_3768c|M.abscessus_ATCC_199 EYRLSTASFGVLPLARHDVVGWHRPLDVDAMNEASRNLLGLRDFAAFCRH
***:.*.:* * : :. * * **: :* *: :::**.:**.***
Mflv_4968|M.gilvum_PYR-GCK RDGATTIRELQRLDCVRDGDRISVHVSADAFCWNMVRSLVGALLAVGEHR
Mvan_1438|M.vanbaalenii_PYR-1 RDGATTIRELQRLDCVRDGDRVEVHVSADAFCWNMVRSLVGALLAVGEHR
MSMEG_1527|M.smegmatis_MC2_155 RAGATTIRDLQRLEWVRNGFYINAYVTADAFCWNMVRSLVGALLKVGEGR
TH_0034|M.thermoresistible__bu REGATTIRDLQRFDWSRHGDLITANVSADAFCWQMVRSLVGAVLKVGEGR
Mb3484c|M.bovis_AF2122/97 REGATTIRDLQRLDWSRAGTLVTAHVTADAFCWSMVRSLVGALLAVGEHR
Rv3455c|M.tuberculosis_H37Rv REGATTIRDLQRLDWSRAGTLVTAHVTADAFCWSMVRSLVGALLAVGEHR
MMAR_1092|M.marinum_M REGATTIRDLQRLDWSRNGHLVTAQVSADAFCWSMVRSLVGALLAVGEHR
MUL_0850|M.ulcerans_Agy99 REGATTIRDLQRLDWSRNGHLVTAQVSADAFCWSMVRSLVGALLAVGEHR
MLBr_01955|M.leprae_Br4923 RHGATTIRELQRLEWSREGYLITAHITADAFCWSMVRSLIGALLAVGDHR
MAV_4395|M.avium_104 RENATTIRDLQRLDWTRDGDLIIARVSADAFCWSMVRSLVGALLAVGENR
MAB_3768c|M.abscessus_ATCC_199 REGATTIRELQRFDWGVDGQRLTAYVSADAFCWSMVRSLVGAVLAVGEGR
* .*****:***:: * : . ::******.*****:**:* **: *
Mflv_4968|M.gilvum_PYR-GCK RDDTWCVQLLHARHRSSDFAAAPARGLTLVGVDYPPDDELAARNLVTRDV
Mvan_1438|M.vanbaalenii_PYR-1 RDAAWCAALLDATGRSSDFAAAPARGLTLVGVDYPPDAQLEARNLVTRDL
MSMEG_1527|M.smegmatis_MC2_155 RSPEWTAELLGATSRSSEFPAAPARGLTLAAVDYPPDDELEARNVVTRDV
TH_0034|M.thermoresistible__bu RDPQWCAALLTATRRSSDFAAAPAQGLTLAGVDYPPDDQLAARIAITRDR
Mb3484c|M.bovis_AF2122/97 RATTWCRELLTATGRSSDFAVAPAHGLTLIQVDYPPDDQLASRNLVTRDV
Rv3455c|M.tuberculosis_H37Rv RATTWCRELLTATGRSSDFAVAPAHGLTLIQVDYPPDDQLASRNLVTRDV
MMAR_1092|M.marinum_M RTPDWCRELLTATARSSDFAAAPAHGLVLAKVDYPPDDQLASRVLITRDL
MUL_0850|M.ulcerans_Agy99 RTPDWCRELLTATARSSDFAAAPAHGLVLAKVDYPPDDQLASRVLITRDL
MLBr_01955|M.leprae_Br4923 RTIGWCGELLTKASRSSDFTTAPAHGLTFIRVDYPPDDELAERAVITRDL
MAV_4395|M.avium_104 RPVSWCRELLSATNRSSDYATAPAHGLTLVGVDYPPDDQLAARNLITRDV
MAB_3768c|M.abscessus_ATCC_199 RPVQWCVDLLNKERRSSDFAAAPARGLTLVGVDYPPDDQMAARVMITRDV
* * ** ***::..***:**.: ****** :: * :***
Mflv_4968|M.gilvum_PYR-GCK RTADEL--
Mvan_1438|M.vanbaalenii_PYR-1 RTADEL--
MSMEG_1527|M.smegmatis_MC2_155 RTIDQSRD
TH_0034|M.thermoresistible__bu REL-----
Mb3484c|M.bovis_AF2122/97 RSG-----
Rv3455c|M.tuberculosis_H37Rv RSG-----
MMAR_1092|M.marinum_M RSRTD---
MUL_0850|M.ulcerans_Agy99 RSRTD---
MLBr_01955|M.leprae_Br4923 RSSF----
MAV_4395|M.avium_104 RSR-----
MAB_3768c|M.abscessus_ATCC_199 RIRPSG--
*