For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVTRIVILGGGPAGYEAALVAATSHPETAQVTVIDCDGIGGAAVLDDCVPSKTFIASTGLRTELRRAPHL GFHIDFDDAKISLPQIHARVKTLAAAQSADITAQLLSMGVQVIAGRGELIDSTPGLARHRIKATAADGST SEHEADVVLVATGASPRILPSAQPDGERILTWRQLYDLDALPDHLIVVGSGVTGAEFVDAYTELGVPVTV VASQDHVLPYEDADAALVLEESFAERGVRLFKNARAASVTRTGAGVLVTMTDGRTVEGSHALMTIGSVPN TSGLGLERVGIQLGRGNYLTVDRVSRTSATGIYAAGDCTGLLPLASVAAMQGRIAMYHALGEGVSPIRLR TVAATVFTRPEIAAVGVPQSVIDAGSVAARTIMLPLRTNARAKMSEMRHGFVKIFCRRSTGVVIGGVVVA PIASELILPIAVAVQNRITVNELAQTLAVYPSLSGSITEAARRLMAHDDLD
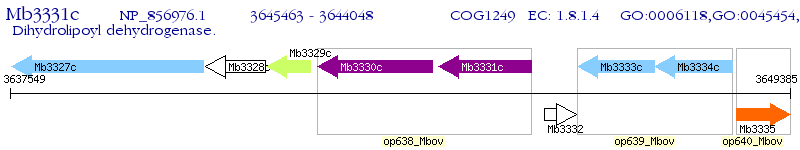
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3331c | lpdA | - | 100% (471) | flavoprotein disulfide reductase |
| M. bovis AF2122 / 97 | Mb0471 | lpd | 9e-44 | 28.30% (470) | dihydrolipoamide dehydrogenase |
| M. bovis AF2122 / 97 | Mb2732 | sthA | 4e-37 | 26.45% (465) | soluble pyridine nucleotide transhydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4822 | - | 0.0 | 76.22% (471) | flavoprotein disulfide reductase |
| M. tuberculosis H37Rv | Rv3303c | lpdA | 0.0 | 99.58% (471) | flavoprotein disulfide reductase |
| M. leprae Br4923 | MLBr_02387 | lpd | 2e-44 | 27.16% (475) | dihydrolipoamide dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3656c | - | 0.0 | 76.65% (471) | flavoprotein disulfide reductase |
| M. marinum M | MMAR_1224 | lpdA | 0.0 | 92.55% (470) | dihydrolipoamide dehydrogenase LpdA |
| M. avium 104 | MAV_4687 | lpdA | 2e-43 | 27.99% (468) | dihydrolipoamide dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1735 | - | 0.0 | 78.98% (471) | flavoprotein disulfide reductase |
| M. thermoresistible (build 8) | TH_1837 | lpdA | 0.0 | 76.22% (471) | PROBABLE DIHYDROLIPOAMIDE DEHYDROGENASE LPDA (LIPOAMIDE |
| M. ulcerans Agy99 | MUL_2668 | lpdA | 0.0 | 92.36% (471) | flavoprotein disulfide reductase |
| M. vanbaalenii PYR-1 | Mvan_1619 | - | 0.0 | 77.49% (471) | flavoprotein disulfide reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4822|M.gilvum_PYR-GCK ------------------------MVTRIVIIGGGPAGYEAALVAAGLGR
Mvan_1619|M.vanbaalenii_PYR-1 MAPLPSGGTCAGTPRPRRWPRFTTVVTRIVIIGGGPAGYEAALVAAGLGR
Mb3331c|M.bovis_AF2122/97 ------------------------MVTRIVILGGGPAGYEAALVAATSHP
Rv3303c|M.tuberculosis_H37Rv ------------------------MVTRIVILGGGPAGYEAALVAATSHP
MMAR_1224|M.marinum_M -------------------------MTRIVILGGGPAGYEAALVAATSHP
MUL_2668|M.ulcerans_Agy99 ------------------------MVTRIVILGGGPAGYEAALVAATSHP
MAB_3656c|M.abscessus_ATCC_199 ------------------------MATRIVIIGGGPAGYEAALVAAAHER
MSMEG_1735|M.smegmatis_MC2_155 ------------------------MVTRIVILGGGPAGYEAALVAAARGP
TH_1837|M.thermoresistible__bu ------------------------VATRIVIIGGGPAGYEAALVAAAKGT
MLBr_02387|M.leprae_Br4923 -----------------------MTHYDVVVLGAGPGGYVAAIRAAQLGL
MAV_4687|M.avium_104 ----------------------MTSHYDVVVLGAGPGGYVAAIRAAQLGL
:*::*.**.** **: **
Mflv_4822|M.gilvum_PYR-GCK ELTQVTVVDSDGLGGACVLYDCVPSKTFIASTGVRTELRRAQGMGYDIGI
Mvan_1619|M.vanbaalenii_PYR-1 DLTQVTVIDSDGLGGACVLYDCVPSKTFIASTGVRTELRRAQGMGFDIGI
Mb3331c|M.bovis_AF2122/97 ETAQVTVIDCDGIGGAAVLDDCVPSKTFIASTGLRTELRRAPHLGFHIDF
Rv3303c|M.tuberculosis_H37Rv ETTQVTVIDCDGIGGAAVLDDCVPSKTFIASTGLRTELRRAPHLGFHIDF
MMAR_1224|M.marinum_M ESAQVTVIDSDGIGGAAVLDDCVPSKTFIASTGLRTELRRAPHLGFLIDF
MUL_2668|M.ulcerans_Agy99 ESAQVTVIDSDGIGGAGVLDDCVPSKTFIASTGLRTELRRAPHLGFLIDF
MAB_3656c|M.abscessus_ATCC_199 STTEVTVIDSDGIGGACVLFDCVPSKTFIASTGVRTDLRRAPNLGFDIDV
MSMEG_1735|M.smegmatis_MC2_155 EVADVTVVDCDGIGGACVLWDCVPSKSFIAATGVRTELRRAPRLGYSIDF
TH_1837|M.thermoresistible__bu EV-EVTIVDSDGIGGACVLWDCVPSKTFIASTGVRTELRRAPNLGYQLEF
MLBr_02387|M.leprae_Br4923 ST---AVVEPKYWGGICLNVGCIPSKVLLHNAELAHIFTKEAKT-FGIS-
MAV_4687|M.avium_104 ST---AIVEPKYWGGVCLNVGCIPSKALLRNAELAHIFTKEAKT-FGIN-
. :::: . ** : .*:*** :: : : : : : :
Mflv_4822|M.gilvum_PYR-GCK DDAPISLPKINNRVKTLAASQSADISSQLLNQRVTIIGGRGELVDDVAGM
Mvan_1619|M.vanbaalenii_PYR-1 DDAPISLPKINNRVKTLAASQSADIGSQLLNQGVTIIGGRGELVDDIAGM
Mb3331c|M.bovis_AF2122/97 DDAKISLPQIHARVKTLAAAQSADITAQLLSMGVQVIAGRGELIDSTPGL
Rv3303c|M.tuberculosis_H37Rv DDAKISLPQIHARVKTLAAAQSADITAQLLSMGVQVIAGRGELIDSTPGL
MMAR_1224|M.marinum_M DEAKISLPKIHARVKELAAAQSADITAQLLSVGVHVIAGKGELLDPAPGL
MUL_2668|M.ulcerans_Agy99 DEAKISLPKIHARVKELAAAQSADITAQLLSVGVHVIAGKGELLDPAPGL
MAB_3656c|M.abscessus_ATCC_199 EHAKISLPQIHSRVKRLASEQSADIAERLRSAGVHLIAGRAELVDHVVGM
MSMEG_1735|M.smegmatis_MC2_155 EQAKVGLPLLNERVKALAAAQSRDIADRLRSEGITLIAGRGELIDDVPGM
TH_1837|M.thermoresistible__bu EDARISLRQINERVKRLAKAQSADIADRLRREGVRLIAGRGELVDAARGM
MLBr_02387|M.leprae_Br4923 GDASFDYGIAYDRSRKVSEGRVAGVHFLMKKNKITEIHGYGRFTDANTLS
MAV_4687|M.avium_104 GEATFDYGAAFDRSRKVAEGRVAGVHFLMKKNKITEIHGYGRFTDPHTLA
.* .. * : :: : .: : : * * ..: *
Mflv_4822|M.gilvum_PYR-GCK AHHRVKVTTHDGKVGVLKADVVLIATGASPRVLPNAVPDGERILTWRHVY
Mvan_1619|M.vanbaalenii_PYR-1 AHHRVKVTTSDGKVGVLKADVVLIATGASPRVLPNAVPDGERILTWRHVY
Mb3331c|M.bovis_AF2122/97 ARHRIKATAADGSTSEHEADVVLVATGASPRILPSAQPDGERILTWRQLY
Rv3303c|M.tuberculosis_H37Rv ARHRIKATAADGSTSEHEADVVLVATGASPRILPSAQPDGERILTWRQLY
MMAR_1224|M.marinum_M R-HRVKATATDGTTSEHDADVVLIATGASPRILPSARPDGERILTWRQLY
MUL_2668|M.ulcerans_Agy99 R-HRVKATAADGTTSEHDADVVLIATGASPRILPSARPDGERILTWRQLY
MAB_3656c|M.abscessus_ATCC_199 AFHRVKATAPDGTSTILEADVVLIATGASPRVLPHARPDGERILTWRQLY
MSMEG_1735|M.smegmatis_MC2_155 AHHRIKVTGHDGTVQQLTADVVLIATGASPRVLPNAEPDGERILNWRQLY
TH_1837|M.thermoresistible__bu AQHRIKVTGNDGSVRELTADVVLIATGGTPRVLPNAVPDGERILNWRQLY
MLBr_02387|M.leprae_Br4923 VELSEGVPETP---LKVTFNNVIIATGSKTRLVPGTLLSTN-VITYEEQI
MAV_4687|M.avium_104 VELNDGGTET------VTFDNAIIATGSSTRLVPGTSLSAN-VVTYEEQI
. : .::***...*::* : . : ::.:..
Mflv_4822|M.gilvum_PYR-GCK DLEELPEHLIIVGSGVTGAEFCNAYTELGVTVTVVASRDQILPHEDSDAA
Mvan_1619|M.vanbaalenii_PYR-1 DLDELPEHLVIVGSGVTGAEFCNAYTELGVTVTVVASRDQILPHEDSDAA
Mb3331c|M.bovis_AF2122/97 DLDALPDHLIVVGSGVTGAEFVDAYTELGVPVTVVASQDHVLPYEDADAA
Rv3303c|M.tuberculosis_H37Rv DLDALPDHLIVVGSGVTGAEFVDAYTELGVPVTVVASQDHVLPYEDADAA
MMAR_1224|M.marinum_M DLDALPQHLIVVGSGVTGAEFVDAYTELGVRVTVVASQDHVLPYEDADAA
MUL_2668|M.ulcerans_Agy99 DLDALPQHLIVVGSGVTGAEFVDAYTELGVRVTVVASQDHVLPYEDADAA
MAB_3656c|M.abscessus_ATCC_199 DLDELPEHLIVVGSGVTGAEFVHAYTELGVQVTVVASRDRVLPHEDEDAA
MSMEG_1735|M.smegmatis_MC2_155 DLDTLPEHLVVVGSGVTGAEFVNAYTELGVDVTVVASRDQILPHEDSDAA
TH_1837|M.thermoresistible__bu DLDTLPEHLVVVGSGVTGAEFVNAYTELGVKVTVVASRDQILPHEDSDAA
MLBr_02387|M.leprae_Br4923 LTRELPDSIVIVGAGAIGIEFGYVLKNYGVDVTIVEFLPRAMPNEDAEVS
MAV_4687|M.avium_104 LSRELPESIIIAGAGAIGMEFGYVLHNYGVEVTIVEFLPRALPNEDADVS
**: :::.*:*. * ** . : ** **:* : :* ** :.:
Mflv_4822|M.gilvum_PYR-GCK AALEQVFNERGVTLIKNARAESVTRTDTGVRVTIADG---RVVEGSHALM
Mvan_1619|M.vanbaalenii_PYR-1 AVLEEVFAERGVTLVKNARADSVTRTETGVKVSMADG---RTVEGSHALM
Mb3331c|M.bovis_AF2122/97 LVLEESFAERGVRLFKNARAASVTRTGAGVLVTMTDG---RTVEGSHALM
Rv3303c|M.tuberculosis_H37Rv LVLEESFAERGVRLFKNARAASVTRTGAGVLVTMTDG---RTVEGSHALM
MMAR_1224|M.marinum_M LVLEESFAERGVRLFKNARAQSVTRTESGVLVTMTDG---RTVEGSHALM
MUL_2668|M.ulcerans_Agy99 LVLEESFAERGVRLFKNARAQSVTRTENGVLVTMTDG---RTVEGSHALM
MAB_3656c|M.abscessus_ATCC_199 LVLEDTLAERGVTLVKNARADSVARTDTGVRVSMADG---RVVDGSHVLM
MSMEG_1735|M.smegmatis_MC2_155 AVLEQVFAQRGVTLVKNARAESVTRTGDGVLVTMTDG---RTVDGSHALM
TH_1837|M.thermoresistible__bu AVLEDVFAERGVTLVKNARADSVKRENGGVRVTMTDG---REVQGSHALI
MLBr_02387|M.leprae_Br4923 KEIEKQFKKMGIKILTGTKVESISDNGSHVLVAVSKDGQFQELKADKVLQ
MAV_4687|M.avium_104 KEIEKQFKKLGVKILTGTKVESISDDGSQVTVVVSKDGNSQELKAAKVLQ
:*. : : *: :...::. *: * * ::.. : :.. :.*
Mflv_4822|M.gilvum_PYR-GCK TVGSVPNTSGLGLERVGVELNPGGYIPVDRVSRTPAPGVYAAGDCTGLLP
Mvan_1619|M.vanbaalenii_PYR-1 TVGSVPNTKNLGLERVGITLNPGGYIPVDRVSRTPAPGVYAAGDCTGLLP
Mb3331c|M.bovis_AF2122/97 TIGSVPNTSGLGLERVGIQLGRGNYLTVDRVSRTSATGIYAAGDCTGLLP
Rv3303c|M.tuberculosis_H37Rv TIGSVPNTSGLGLERVGIQLGRGNYLTVDRVSRTLATGIYAAGDCTGLLP
MMAR_1224|M.marinum_M TIGSIPNTSGLGLERVGIELGPGNYLNVDRVSRTSVPGIYAAGDCTGLLP
MUL_2668|M.ulcerans_Agy99 TIGSIPNTSGLGLERVGIELGPGNYLNVDRVSRTSVPGVYAAGDCTGLLP
MAB_3656c|M.abscessus_ATCC_199 TVGSIPNTAGLGLERVGITLGPGGYLTVDRVSRTSVAGIYAAGDCTGLLP
MSMEG_1735|M.smegmatis_MC2_155 TVGSVPNTSGLGLERVGIELGAGGYLTVDRVSRTQVPGIYAAGDCTGLLP
TH_1837|M.thermoresistible__bu TVGSVPNTANLGLERVGIELGPGGYLPVDRVSRTTVPGIYAAGDCTGLLP
MLBr_02387|M.leprae_Br4923 AIGFAPNVDGYGLDKVGVALTADKAVDIDDYMQTNVSHIYAIGDVTGKLQ
MAV_4687|M.avium_104 AIGFAPNVEGYGLEAAGVALTDRKAIGITDYMRTNIEHIYAIGDVTGKLQ
::* **. . **: .*: * : : :* :** ** ** *
Mflv_4822|M.gilvum_PYR-GCK LASVAAMQGRIAMYHALG-EGVAPIRLRTVASATFTRPEIAAVGIPQSAI
Mvan_1619|M.vanbaalenii_PYR-1 LASVAAMQGRIAMYHALG-EGVSPIRLRTVASATFTRPEIAAVGIPQSAI
Mb3331c|M.bovis_AF2122/97 LASVAAMQGRIAMYHALG-EGVSPIRLRTVAATVFTRPEIAAVGVPQSVI
Rv3303c|M.tuberculosis_H37Rv LASVAAMQGRIAMYHALG-EGVSPIRLRTVAATVFTRPEIAAVGVPQSVI
MMAR_1224|M.marinum_M LASVAAMQGRIAMYHALG-EGVSPIRLRTVAATVFTRPEIAAVGVPQSAI
MUL_2668|M.ulcerans_Agy99 LASVAAMQGRIAMYHALG-EGVSPIRLRTVAATVFTRPEIAAVGVPQSAI
MAB_3656c|M.abscessus_ATCC_199 LASVAAMQGRIAMYHALG-DAVQPIRLKTVAAAVFTRPEIAAVGVSQAAI
MSMEG_1735|M.smegmatis_MC2_155 LASVAAMQGRIAMYHALG-EAVAPIRLRTVAAAVFTRPEIAAVGVPQAKI
TH_1837|M.thermoresistible__bu LASVAAMQGRIAMYHALG-EALNPIRLRTVAAAVFTRPEIAAVGVPQAKI
MLBr_02387|M.leprae_Br4923 LAHVAEAQGVVAAEAIAGAETLALSDYRMMPRATFCQPNVASFGLTEQQA
MAV_4687|M.avium_104 LAHVAEAQGVVAAETIAGAETLALGDYRMMPRATFCQPNVASFGLTEQQA
** ** ** :* * : : : :. :.* :*::*:.*:.:
Mflv_4822|M.gilvum_PYR-GCK DDGSVPARTLMLPLSTNARAKMSLLRHGFVKIFCRPATGVVIGGVVVAPI
Mvan_1619|M.vanbaalenii_PYR-1 DDGSVPARTLMLPLSTNARAKMSLLRHGFVKIFCRPATGVVIGGVVVAPI
Mb3331c|M.bovis_AF2122/97 DAGSVAARTIMLPLRTNARAKMSEMRHGFVKIFCRRSTGVVIGGVVVAPI
Rv3303c|M.tuberculosis_H37Rv DAGSVAARTIMLPLRTNARAKMSEMRHGFVKIFCRRSTGVVIGGVVVAPI
MMAR_1224|M.marinum_M DNGSVLARTIMLPLRTNARAKMSEVRHGFVKVFCRRSTGVVIGGVVVAPI
MUL_2668|M.ulcerans_Agy99 DNGSVLARTIMLPLRTNARAKMSEVRHGFVKVFCRRSTGVVIGGVVVAPI
MAB_3656c|M.abscessus_ATCC_199 DNGDVPARTVTLPLHTNPRAKMSGLRRGFVKIFCRPATGVVIGGVVVAAN
MSMEG_1735|M.smegmatis_MC2_155 DDGSVPARTLTLPLATNARAKMSSLTHGFVKIFCRPATGVVIGGVVVAPI
TH_1837|M.thermoresistible__bu DDGSVPARTIMLPLNTNARAKMSSLRRGFVKIFCRPMTGVVIGGVVVAPI
MLBr_02387|M.leprae_Br4923 RDGGYDVVVAKFPFTANAKAHGMGDPSGFVKLVADAKYGELLGGHMIGHN
MAV_4687|M.avium_104 RDEGHDVVVAKFPFTANGKAHGVGDPSGFVKLIADAKYGELLGGHLVGHD
. . . :*: :* :*: ****:.. * ::** ::.
Mflv_4822|M.gilvum_PYR-GCK ASELILPIALAVQNRISVTDLAQTLSVYPSLSGSIVEAARRLMAHDDLD-
Mvan_1619|M.vanbaalenii_PYR-1 ASELILPIALAVQNRISVSDLAQTLSVYPSLSGSIVEAARRLMAHDDLD-
Mb3331c|M.bovis_AF2122/97 ASELILPIAVAVQNRITVNELAQTLAVYPSLSGSITEAARRLMAHDDLD-
Rv3303c|M.tuberculosis_H37Rv ASELILPIAVAVQNRITVNELAQTLAVYPSLSGSITEAARRLMAHDDLDC
MMAR_1224|M.marinum_M ASELILPIAVAVQNRITVNELAQTLAVYPSLSGSITEAARRLMAHDDLD-
MUL_2668|M.ulcerans_Agy99 ASELILPIAVAVQNRITVNELAQTLAVYPSLSGSITEAARRLMAHDDLD-
MAB_3656c|M.abscessus_ATCC_199 ASELILPIAIAVQNLLSVNDLAQTLSVYPSLSGSVTEAARQLIAHDDLD-
MSMEG_1735|M.smegmatis_MC2_155 ASELILPIALAVQNGIPVSELAQTFSVYPSLSGSITEAARQLMQHDDLD-
TH_1837|M.thermoresistible__bu ASELILPIALAVQNGNTVADLAQTFSVYPSLSGSITEAARQLIAHDDLD-
MLBr_02387|M.leprae_Br4923 VSELLPELTLAQKWDLTATELVRNVHTHPTLSEALQECFHGLIGHMINF-
MAV_4687|M.avium_104 VSELLPELTLAQKWDLTATELARNVHTHPTMSEALQECFHGLTGHMINF-
.***: :::* : .. :*.:.. .:*::* :: *. : * *
Mflv_4822|M.gilvum_PYR-GCK ---------------------
Mvan_1619|M.vanbaalenii_PYR-1 ---------------------
Mb3331c|M.bovis_AF2122/97 ---------------------
Rv3303c|M.tuberculosis_H37Rv TAAQDAAEQLALVPHHLPTSN
MMAR_1224|M.marinum_M ---------------------
MUL_2668|M.ulcerans_Agy99 ---------------------
MAB_3656c|M.abscessus_ATCC_199 ---------------------
MSMEG_1735|M.smegmatis_MC2_155 ---------------------
TH_1837|M.thermoresistible__bu ---------------------
MLBr_02387|M.leprae_Br4923 ---------------------
MAV_4687|M.avium_104 ---------------------