For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VASHAGSRIARISKVLVANRGEIAVRVIRAARDAGLPSVAVYAEPDAESPHVRLADEAFALGGQTSAESY LDFAKILDAAAKSGANAIHPGYGFLAENADFAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPL VPGTPDPVKGADEVVAFAEEYGLPIAIKAAHGGGGKGMKVARTIDEIPELYESAVREATAAFGRGECYVE RYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRYQKLVEEAPAPFLTDFQRKEIHDSAKRICKEAHYHG AGTVEYLVGQDGLISFLEVNTRLQVEHPVTEETAGIDLVLQQFRIANGEKLDITEDPTPRGHAIEFRING EDAGRNFLPAPGPVTKFHPPSGPGVRVDSGVETGSVIGGQFDSMLAKLIVHGADRAEALARARRALNEFG VEGLATVIPFHRAVVSDPAFIGDANGFSVHTRWIETEWNNTIEPFTDGEPLDEDARPRQKVVVEIDGRRV EVSLPADLALSNGGGCDPVGVIRRKPKPRKRGAHTGAAASGDAVTAPMQGTVVKFAVEEGQEVVAGDLVV VLEAMKMENPVTAHKDGTITGLAVEAGAAITQGTVLAEIK
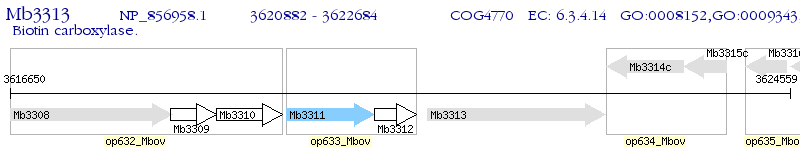
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3313 | accA3 | - | 100% (600) | bifunctional acetyl-/propionyl-coenzyme A carboxylase subunit alpha |
| M. bovis AF2122 / 97 | Mb2529c | accA1 | e-110 | 50.00% (426) | acetyl-/propionyl-coenzyme A carboxylase subunit alpha |
| M. bovis AF2122 / 97 | Mb0998c | accA2 | 7e-95 | 36.12% (670) | acetyl-/propionyl-coenzyme A carboxylase subunit alpha |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4784 | - | 0.0 | 85.36% (601) | carbamoyl-phosphate synthase L chain, ATP-binding |
| M. tuberculosis H37Rv | Rv3285 | accA3 | 0.0 | 100.00% (600) | bifunctional acetyl-/propionyl-coenzyme A carboxylase |
| M. leprae Br4923 | MLBr_00726 | bccA | 0.0 | 88.85% (601) | acetyl/propionyl CoA carboxylase alpha subunit |
| M. abscessus ATCC 19977 | MAB_3643 | - | 0.0 | 83.55% (602) | bifunctional protein acetyl-/propionyl-CoA carboxylase (alpha |
| M. marinum M | MMAR_1251 | accA3 | 0.0 | 91.85% (601) | bifunctional protein acetyl-/propionyl-coenzyme A |
| M. avium 104 | MAV_4255 | - | 0.0 | 90.80% (587) | acetyl-/propionyl-coenzyme A carboxylase alpha chain |
| M. smegmatis MC2 155 | MSMEG_1807 | - | 0.0 | 85.86% (601) | acetyl-/propionyl-coenzyme A carboxylase alpha chain |
| M. thermoresistible (build 8) | TH_0244 | accA3 | 0.0 | 87.85% (601) | PROBABLE BIFUNCTIONAL PROTEIN ACETYL-/PROPIONYL-COENZYME A |
| M. ulcerans Agy99 | MUL_2637 | accA3 | 0.0 | 91.35% (601) | bifunctional protein acetyl-/propionyl-coenzyme a |
| M. vanbaalenii PYR-1 | Mvan_1664 | - | 0.0 | 84.69% (601) | carbamoyl-phosphate synthase L chain, ATP-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4784|M.gilvum_PYR-GCK MASHASS---KISKVLVANRGEIAVRVIRAAKDAGLQSVAVYAEPDADAP
Mvan_1664|M.vanbaalenii_PYR-1 MASHASS---KISKVLVANRGEIAVRVIRAAKDAGLQSVAVYAEPDADAP
MSMEG_1807|M.smegmatis_MC2_155 MANHASS---KISKVLVANRGEIAVRVIRAAKDAGLASVAVYAEPDADAP
Mb3313|M.bovis_AF2122/97 MASHAGSRIARISKVLVANRGEIAVRVIRAARDAGLPSVAVYAEPDAESP
Rv3285|M.tuberculosis_H37Rv MASHAGSRIARISKVLVANRGEIAVRVIRAARDAGLPSVAVYAEPDAESP
MMAR_1251|M.marinum_M MASHASS---RISKVLVANRGEIAVRVIRAARDAGLASVAVYAEPDAEAP
MUL_2637|M.ulcerans_Agy99 MASHASS---RISKVLVANRGEIAVRVIRAARDAGLASVAVYAEPDAEAP
MLBr_00726|M.leprae_Br4923 MASHASS---RIAKVLVANRGEIAVRVIRAARDARLPSVAVYAEPDAEAP
MAV_4255|M.avium_104 --------------MLVANRGEIAVRVIRAARDAGLSSVAVYAEPDADAP
TH_0244|M.thermoresistible__bu VASHASS---KISKVLVANRGEIAVRVIRAARDAGIESVAVYAEPDADAP
MAB_3643|M.abscessus_ATCC_1997 MPNHASS---KISKVLVANRGEIAVRVIRAAKDAGLGSVAIYAEPDADAP
:****************:** : ***:******::*
Mflv_4784|M.gilvum_PYR-GCK HVRLADEAFALGGQTSAESYLVFEKLLDAAAKSGANAIHPGYGFLSENAD
Mvan_1664|M.vanbaalenii_PYR-1 HVRLADEAFALGGQTSAESYLVFEKLLDAAAKSGANAIHPGYGFLSENAD
MSMEG_1807|M.smegmatis_MC2_155 HVRLADEAFALGGQTSAESYLVFEKILDAAEKSGANAIHPGYGFLSENAD
Mb3313|M.bovis_AF2122/97 HVRLADEAFALGGQTSAESYLDFAKILDAAAKSGANAIHPGYGFLAENAD
Rv3285|M.tuberculosis_H37Rv HVRLADEAFALGGQTSAESYLDFAKILDAAAKSGANAIHPGYGFLAENAD
MMAR_1251|M.marinum_M HVSLADEAFALGGQTSAESYLDFEKLLDAAAKSGANAIHPGYGFLSENAD
MUL_2637|M.ulcerans_Agy99 HVSLADEAFALGGQTSAESYLDFEKLLDAATKSGANAIHPGYGFLSENAD
MLBr_00726|M.leprae_Br4923 HVRLADEAFALGGHTSAESYLDFGKILDAAAKSGANAIHPGYGFLAENAD
MAV_4255|M.avium_104 HVRLADEAFALGGQTSAESYLDFGKLLDAAAKSGANAIHPGYGFLSENAD
TH_0244|M.thermoresistible__bu HVRLADEAFALGGQTSAESYLVFEKILDAAAKSGANAIHPGYGFLSENAD
MAB_3643|M.abscessus_ATCC_1997 HVHLADEAFGIGGNTAAESYLDFGKILEAAEKSGANAIHPGYGFLSENAD
** ******.:**:*:***** * *:*:** **************:****
Mflv_4784|M.gilvum_PYR-GCK FAQAVLDAGLIWIGPSPQSIRDLGDKVTARHIAARANAPLVPGTPDPVKD
Mvan_1664|M.vanbaalenii_PYR-1 FAQAVLDAGLIWIGPSPQSIRDLGDKVTARHIAARAEAPLVPGTPDPVKD
MSMEG_1807|M.smegmatis_MC2_155 FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAKAPLVPGTPDPVKD
Mb3313|M.bovis_AF2122/97 FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPLVPGTPDPVKG
Rv3285|M.tuberculosis_H37Rv FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPLVPGTPDPVKG
MMAR_1251|M.marinum_M FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAKAPLVPGTPDPVKN
MUL_2637|M.ulcerans_Agy99 FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAKAPLVPGTPDPVKN
MLBr_00726|M.leprae_Br4923 FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPLVPGTPDPVKN
MAV_4255|M.avium_104 FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPLVPGTPDPVKD
TH_0244|M.thermoresistible__bu FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAKAPLVPGTADPVKD
MAB_3643|M.abscessus_ATCC_1997 FAQAVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPLVPGTPDPVKD
*****:******************************:*******.****.
Mflv_4784|M.gilvum_PYR-GCK ADEVVAFAKEYGVPVAIKAAFGGGGRGMKVARTIEEIPELFDSATREAVA
Mvan_1664|M.vanbaalenii_PYR-1 ADEVVAFAKEYGVPVAIKAAFGGGGRGMKVARTIEEIPELFDSATREAVA
MSMEG_1807|M.smegmatis_MC2_155 ADEVVAFAKEHGVPVAIKAAFGGGGRGMKVARTLEEIPELFESATREAIA
Mb3313|M.bovis_AF2122/97 ADEVVAFAEEYGLPIAIKAAHGGGGKGMKVARTIDEIPELYESAVREATA
Rv3285|M.tuberculosis_H37Rv ADEVVAFAEEYGLPIAIKAAHGGGGKGMKVARTIDEIPELYESAVREATA
MMAR_1251|M.marinum_M AEEVVAFAQEYGLPIAIKAAFGGGGRGMKVARTMEEIPELYESAVREATA
MUL_2637|M.ulcerans_Agy99 AEEVVAFAQEYGLPIAIKAAFGGGGRGMKVARTVEEIPELYESAVREATA
MLBr_00726|M.leprae_Br4923 ADEVVAFAKEHGVPIAIKAAFGGGGKGMKVARTLEEISELYESAVREATV
MAV_4255|M.avium_104 ADEVVAFAKEYGVPIAIKAAFGGGGKGMKVARTIEEIPELYESAVREAVA
TH_0244|M.thermoresistible__bu ADEVVAFAKEHGVPIAIKAAFGGGGRGMKVARTIEEIPDLYESAVREAVA
MAB_3643|M.abscessus_ATCC_1997 ADEVVAFAKEHGLPIAIKAAFGGGGRGMKVARTLEEVPELFESATREAVA
*:******:*:*:*:*****.****:*******::*:.:*::**.*** .
Mflv_4784|M.gilvum_PYR-GCK AFGRGECFVERYLDKPRHVEAQVIADTHGNVVVAGTRDCSLQRRFQKLVE
Mvan_1664|M.vanbaalenii_PYR-1 AFGRGECFVERYLDKPRHVEAQVIADMHGNVIVAGTRDCSLQRRFQKLVE
MSMEG_1807|M.smegmatis_MC2_155 AFGRGECFVERYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRFQKLVE
Mb3313|M.bovis_AF2122/97 AFGRGECYVERYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRYQKLVE
Rv3285|M.tuberculosis_H37Rv AFGRGECYVERYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRYQKLVE
MMAR_1251|M.marinum_M AFGRGECFVERYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRYQKLVE
MUL_2637|M.ulcerans_Agy99 AFGRGECFVERYLDRPRHVEAQVIADQHGNVVVAGTRDCSLQRRYQKLVE
MLBr_00726|M.leprae_Br4923 AFGRGECFVERYLDKPRHVEAQVIADQHGNIVVAGTRDCSLQRRFQKLVE
MAV_4255|M.avium_104 AFGRGECFVERYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRFQKLVE
TH_0244|M.thermoresistible__bu AFGRGECFVERYLDKPRHVEAQVLADQHGNVVVAGTRDCSLQRRFQKLVE
MAB_3643|M.abscessus_ATCC_1997 AFGRGECFVERYLDKPRHVEAQVIADQHGNVVVAGTRDCSLQRRFQKLVE
*******:******:********:** ***::************:*****
Mflv_4784|M.gilvum_PYR-GCK EAPAPFLTDAQRKEIHESAKRICKEAGYYGAGTVEYLVGQDGLISFLEVN
Mvan_1664|M.vanbaalenii_PYR-1 EAPAPFLTDAQRKEIHESAKRICKEAGYYGAGTVEYLVGQDGLISFLEVN
MSMEG_1807|M.smegmatis_MC2_155 EAPAPFLTDAQRKEIHESAKRICKEAGYYGAGTVEYLVGQDGLISFLEVN
Mb3313|M.bovis_AF2122/97 EAPAPFLTDFQRKEIHDSAKRICKEAHYHGAGTVEYLVGQDGLISFLEVN
Rv3285|M.tuberculosis_H37Rv EAPAPFLTDFQRKEIHDSAKRICKEAHYHGAGTVEYLVGQDGLISFLEVN
MMAR_1251|M.marinum_M EAPAPFLSDAQRKEIHESAKRICKEAHYYGAGTVEYLVGQDGLISFLEVN
MUL_2637|M.ulcerans_Agy99 EAPAPFLSDAQRKEIHESAKRICKEAHYYGAGTVEYLVGQDGLISFLEVN
MLBr_00726|M.leprae_Br4923 EAPAPFLTDAQRKEIHESAKRICKEAHYYGAGTVEYLVGQDGLISFLEVN
MAV_4255|M.avium_104 EAPAPFLSDAQRKEIHESAKRICKEAHYYGAGTVEYLVGQDGLISFLEVN
TH_0244|M.thermoresistible__bu EAPAPFLTDAQRKEIHESAKRICKEAGYYGAGTVEYLVGQDGMISFLEVN
MAB_3643|M.abscessus_ATCC_1997 EAPAPFLTDAQRKEIHESAKRICKEAGYYGAGTVEYLVGQDGLISFLEVN
*******:* ******:********* *:*************:*******
Mflv_4784|M.gilvum_PYR-GCK TRLQVEHPVTEETSGIDLVLQQFKIANGEALDITEDPTPRGHAFEFRING
Mvan_1664|M.vanbaalenii_PYR-1 TRLQVEHPVTEETSGIDLVLQQFKIANGEALDITEDPTPRGHSFEFRING
MSMEG_1807|M.smegmatis_MC2_155 TRLQVEHPVTEETSGIDLVRQQFKIANGEPLDITEDPTPRGHSFEFRING
Mb3313|M.bovis_AF2122/97 TRLQVEHPVTEETAGIDLVLQQFRIANGEKLDITEDPTPRGHAIEFRING
Rv3285|M.tuberculosis_H37Rv TRLQVEHPVTEETAGIDLVLQQFRIANGEKLDITEDPTPRGHAIEFRING
MMAR_1251|M.marinum_M TRLQVEHPVTEETAGIDLVREQFRIANGEKLDLTEDPTPRGHAIEFRING
MUL_2637|M.ulcerans_Agy99 TRLQVEHPVTEETAGIDLVREQFRIANGEKLDLTEDPTPRGHAIEFRING
MLBr_00726|M.leprae_Br4923 TRLQVEHPVTEETTGIDLVLQQFKIANGEKLELIKDPIPCGHAIEFRING
MAV_4255|M.avium_104 TRLQVEHPVTEETAGIDLVLQQFKIANGDKLDITEDPEPRGHAIEFRING
TH_0244|M.thermoresistible__bu TRLQVEHPVTEETAGIDLVLQQFKIANGEKLDITEDPEPRGHSFEFRING
MAB_3643|M.abscessus_ATCC_1997 TRLQVEHPVTEETAGVDLVLEQFKIANGEALQFTEDPEPRGHSIEFRING
*************:*:*** :**:****: *:: :** * **::******
Mflv_4784|M.gilvum_PYR-GCK EDAGRGFLPAPGPVTKFEPPTGPGVRMDSGVESGSVIGGQFDSMLAKLIV
Mvan_1664|M.vanbaalenii_PYR-1 EDAGRGFLPAPGPVSKFEAPTGPGVRMDSGVETGSVIGGQFDSMLAKLIV
MSMEG_1807|M.smegmatis_MC2_155 EDAGRGFLPAPGPVTKFVAPTGPGVRMDSGVETGSVIGGQFDSMLAKLIV
Mb3313|M.bovis_AF2122/97 EDAGRNFLPAPGPVTKFHPPSGPGVRVDSGVETGSVIGGQFDSMLAKLIV
Rv3285|M.tuberculosis_H37Rv EDAGRNFLPAPGPVTKFHPPSGPGVRVDSGVETGSVIGGQFDSMLAKLIV
MMAR_1251|M.marinum_M EDAGRGFLPAPGPVTKFHPPAGPGVRMDSGVETGSVIGGQFDSMLAKLIV
MUL_2637|M.ulcerans_Agy99 EDAGRGFLPAPGPVTKFHPPAGPGVRMDSGVETGSVIGGQFDSMLAKLIV
MLBr_00726|M.leprae_Br4923 EDAGRNFLPSPGPVSKFHPPTGPGVRLDSGVETGSVIGGQFDSMLAKLIV
MAV_4255|M.avium_104 EDAGRNFLPAPGPVTRYDIPTGPGVRLDSGVEAGSVIGGQFDSMLSKLIV
TH_0244|M.thermoresistible__bu EDAGRGFLPAPGPVTKFAPPSGPGVRLDSGVETGSVIGGQFDSMLAKLIV
MAB_3643|M.abscessus_ATCC_1997 EDAGRNFLPAPGPVKVYDTPTGPGVRLDSGVQAGSVIGGQFDSMLAKLIV
*****.***:****. : *:*****:****::************:****
Mflv_4784|M.gilvum_PYR-GCK VGATREEALARSRRALAEFNVEGLATVIPFHRAVTADPAFIGDGETFTVH
Mvan_1664|M.vanbaalenii_PYR-1 TGATRDEALARSRRALAEFNVEGLATVIPFHRAVVADPAFIGDGEKFDVH
MSMEG_1807|M.smegmatis_MC2_155 TGATREEALERSRRALAEFTVEGLATVIPFHRAVVSDPAFIGDGEKFDVH
Mb3313|M.bovis_AF2122/97 HGADRAEALARARRALNEFGVEGLATVIPFHRAVVSDPAFIGDANGFSVH
Rv3285|M.tuberculosis_H37Rv HGADRAEALARARRALNEFGVEGLATVIPFHRAVVSDPAFIGDANGFSVH
MMAR_1251|M.marinum_M YGADRTEALERARRALAEFQVEGLATVIPFHRAVVSDPAFIGDEHGFSVH
MUL_2637|M.ulcerans_Agy99 YGANRTEALERARRALAEFQVEGLATVIPFHRAVVSDPAFIGDEHGFSVH
MLBr_00726|M.leprae_Br4923 HGATRQEALARARRALDEFEVEGLATVIPFHRAVVSDPALIGDNNSFSVH
MAV_4255|M.avium_104 YGADRQQALARARRALDEFHVEGLATVIPFHRAVVRDPAFIGDDNGFAVH
TH_0244|M.thermoresistible__bu TGATREEALARARRALNEFEIEGLATVLPFHRAVVDDPAFIGDENGFSVH
MAB_3643|M.abscessus_ATCC_1997 TGRDRNEALARSRRALAEFNVEGLATVIPFHRAVVSDPAFIGDEDGFTVH
* * :** *:**** ** :******:******. ***:*** . * **
Mflv_4784|M.gilvum_PYR-GCK TRWIETEWDNTVEPFTGG-DPIDEEDTIPRQTVVVEVGGRRLEVSLPGDL
Mvan_1664|M.vanbaalenii_PYR-1 TRWIETEWDNTVEPFTGG-DPIDEEDTIPRQTVVVEVGGRRLEVSLPGDL
MSMEG_1807|M.smegmatis_MC2_155 TRWIETEWNNTVEPFTGG-DPIEEEDTVPRQTVVVEVGGRRLEVSLPGDL
Mb3313|M.bovis_AF2122/97 TRWIETEWNNTIEPFTDG-EPLD-EDARPRQKVVVEIDGRRVEVSLPADL
Rv3285|M.tuberculosis_H37Rv TRWIETEWNNTIEPFTDG-EPLD-EDARPRQKVVVEIDGRRVEVSLPADL
MMAR_1251|M.marinum_M TRWIETEWNNTIEPFTAG-EPLDDEDFRPRQTVVVEIDGRRVEVSLPADL
MUL_2637|M.ulcerans_Agy99 TRWIETEWNNTIEPFTAG-EPLDDEDFRPRQTVVVEIDGRRVEVSLPADL
MLBr_00726|M.leprae_Br4923 TRWIETEWNNTIEPFIDN-QPLDEEDTRPQQTVIVEVDGRRLEVSLPADL
MAV_4255|M.avium_104 TRWIETEWNNTVEPFTGG-EPLDDEDARPRQKVVVEVGGRRLEVSLPGDL
TH_0244|M.thermoresistible__bu TRWIETEWNNTVEPFTGG-GEVDAEEPEPRQKVVVEVDGRRLEVSLPGNL
MAB_3643|M.abscessus_ATCC_1997 TRWIETEWNNTIEPFTAGGEAADEDEALPRQKLVVEVGGRRLEVSLPGDI
********:**:*** . : :: *:*.::**:.***:*****.::
Mflv_4784|M.gilvum_PYR-GCK AVGGG--APAHGAIRKKPKPRKRGGGGGAAASGDAVTAPMQGTVVKVAVE
Mvan_1664|M.vanbaalenii_PYR-1 ALGGGG-APAHGALRKKPKPRKRGGGGATAASGDAVTAPMQGTVVKVAVE
MSMEG_1807|M.smegmatis_MC2_155 AIGGGGGAAAPGVVRKKPKPRKRGGGGAKAASGDAVTAPMQGTVVKVAVE
Mb3313|M.bovis_AF2122/97 ALSNGGGCDPVGVIRRKPKPRKRGAHTGAAASGDAVTAPMQGTVVKFAVE
Rv3285|M.tuberculosis_H37Rv ALSNGGGCDPVGVIRRKPKPRKRGAHTGAAASGDAVTAPMQGTVVKFAVE
MMAR_1251|M.marinum_M AMSNGSGAD-AGVIRRKPKPRKRGAHTGAAASGDAVTAPMQGTVVKVAVE
MUL_2637|M.ulcerans_Agy99 AMSNGSGAD-AGVIRRKPKPRKRGAHTGAAASGDAVTAPMQGTVVKVAVE
MLBr_00726|M.leprae_Br4923 ALANPAGCNPAGVIRKKPKPRKRGGHTGAATSGDAVTAPMQGTVVKVAVA
MAV_4255|M.avium_104 ALSNGGSADPAGVIRKKPKPRKRGGHGGATASGDAVTAPMQGTVVKVAVE
TH_0244|M.thermoresistible__bu ALGNGTGGA-PGAVRKKPKPRKRGAHGGAAVSGDAVTAPMQGTVVKVAVE
MAB_3643|M.abscessus_ATCC_1997 SLGGGGGAA-NGVVRKKPKARKRGGHGGGAATGDSVTAPMQGTVVKVAVE
::.. *.:*:***.****. . :.:**:***********.**
Mflv_4784|M.gilvum_PYR-GCK EGQTVAAGDLVVVLEAMKMENPVTAHKDGVVTGLSVEPGAAITQGTVIAE
Mvan_1664|M.vanbaalenii_PYR-1 EGQTVAAGDLVVVLEAMKMENPVTAHKDGVVTGLSVEAGAAITQGTVVCE
MSMEG_1807|M.smegmatis_MC2_155 EGQEVSAGDLVVVLEAMKMENPVTAHKDGTITGLAVEAGAAITQGTVIAE
Mb3313|M.bovis_AF2122/97 EGQEVVAGDLVVVLEAMKMENPVTAHKDGTITGLAVEAGAAITQGTVLAE
Rv3285|M.tuberculosis_H37Rv EGQEVVAGDLVVVLEAMKMENPVTAHKDGTITGLAVEAGAAITQGTVLAE
MMAR_1251|M.marinum_M EGQEVVVGELVVVLEAMKMENPVTAHKDGVITGLAVEAGAAITQGTVLAE
MUL_2637|M.ulcerans_Agy99 EGQEVVVGELVVVLEAMKMENPVTAHKDGVITGLAVEAGAAITQGTVLAE
MLBr_00726|M.leprae_Br4923 EGQTVMTGDLVVVLEAMKMENPVTAHKDGIITGLAVEAGTAITQGTVLAE
MAV_4255|M.avium_104 EGQEVSAGDLVVVLEAMKMENPVTAHKDGVITGLAVEAGAAITQGTVLAE
TH_0244|M.thermoresistible__bu EGQEVSAGDLIVVLEAMKMENPVTAHKDGKITGLAVEAGAAITQGTVLAE
MAB_3643|M.abscessus_ATCC_1997 EGQEVEAGELIVVLEAMKMENPVTAHKAGTVTGLSVEAGAAITQGTVIAE
*** * .*:*:**************** * :***:**.*:*******:.*
Mflv_4784|M.gilvum_PYR-GCK LKDAE
Mvan_1664|M.vanbaalenii_PYR-1 LKSAE
MSMEG_1807|M.smegmatis_MC2_155 IK---
Mb3313|M.bovis_AF2122/97 IK---
Rv3285|M.tuberculosis_H37Rv IK---
MMAR_1251|M.marinum_M IK---
MUL_2637|M.ulcerans_Agy99 IK---
MLBr_00726|M.leprae_Br4923 IK---
MAV_4255|M.avium_104 IK---
TH_0244|M.thermoresistible__bu IK---
MAB_3643|M.abscessus_ATCC_1997 IK---
:*