For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTETTDSPSERQPGPAEPELSSRDPDIAGQVFDAAPFDAAPDADSEGDSKAAKTDEPRPAKRSTLREFAV LAVIAVVLYYVMLTFVARPYLIPSESMEPTLHGCSTCVGDRIMVDKLSYRFGSPQPGDVIVFRGPPSWNV GYKSIRSHNVAVRWVQNALSFIGFVPPDENDLVKRVIAVGGQTVQCRSDTGLTVNGRPLKEPYLDPATMM ADPSIYPCLGSEFGPVTVPPGRVWVMGDNRTHSADSRAHCPLLCTNDPLPGTVPVANVIGKARLIVWPPS RWGVVRSVNPQQGR
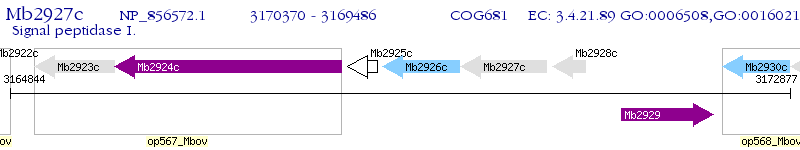
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2927c | lepB | - | 100% (294) | signal peptidase I LepB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4169 | - | 1e-111 | 63.95% (294) | signal peptidase I |
| M. tuberculosis H37Rv | Rv2903c | lepB | 1e-175 | 99.66% (294) | signal peptidase I LepB |
| M. leprae Br4923 | MLBr_01612 | - | 1e-114 | 69.77% (301) | putative ignal peptidase I |
| M. abscessus ATCC 19977 | MAB_3223c | - | 3e-93 | 63.53% (266) | signal peptidase I LepB |
| M. marinum M | MMAR_1805 | lepB | 1e-138 | 79.39% (296) | signal peptidase I LepB |
| M. avium 104 | MAV_3758 | lepB | 1e-124 | 72.04% (304) | signal peptidase I |
| M. smegmatis MC2 155 | MSMEG_2441 | lepB | 1e-112 | 71.33% (279) | signal peptidase I |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2053 | lepB | 1e-138 | 79.39% (296) | signal peptidase I LepB |
| M. vanbaalenii PYR-1 | Mvan_2193 | - | 1e-113 | 67.34% (297) | signal peptidase I |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4169|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2193|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2441|M.smegmatis_MC2_155 --------------------------------------------------
Mb2927c|M.bovis_AF2122/97 --------------------------------------------------
Rv2903c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1805|M.marinum_M --------------------------------------------------
MUL_2053|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01612|M.leprae_Br4923 --------------------------------------------------
MAV_3758|M.avium_104 --------------------------------------------------
MAB_3223c|M.abscessus_ATCC_199 MSYDVYFVRQHPPQDFEHVPENEHPLKESSDLRSRLVADMNAPEVFEGSD
Mflv_4169|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2193|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2441|M.smegmatis_MC2_155 --------------------------------------------------
Mb2927c|M.bovis_AF2122/97 --------------------------------------------------
Rv2903c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1805|M.marinum_M --------------------------------------------------
MUL_2053|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01612|M.leprae_Br4923 --------------------------------------------------
MAV_3758|M.avium_104 --------------------------------------------------
MAB_3223c|M.abscessus_ATCC_199 GTWELSDSVTTAQVNYAPDGVGMGVSYGSATSDEFDSRIEHLMRLAKIIE
Mflv_4169|M.gilvum_PYR-GCK ----MTPSPEPSGPSGPSGIPEDSPTGR-------------------TLE
Mvan_2193|M.vanbaalenii_PYR-1 ----MTSASDPSGHSAPS-DPDNSSAER-------------------TLD
MSMEG_2441|M.smegmatis_MC2_155 ----MTGSTDSTDASSEDALDHDSAGDS-------------------AAN
Mb2927c|M.bovis_AF2122/97 ----MTETTDSPSERQP--GPAEPELSSRDPDIAGQ---VFDAAPFDAAP
Rv2903c|M.tuberculosis_H37Rv ----MTETTDSPSERQP--GPAEPELSSRDPDIAGQ---VFDAAPFDAAP
MMAR_1805|M.marinum_M ----MTETTDSPSEREPDAGQSEAPLAARGADAPG------------AAA
MUL_2053|M.ulcerans_Agy99 ----MTETTDSPSEREPDAGQSEAPLTARGADAPG------------AAA
MLBr_01612|M.leprae_Br4923 ----MTETTDSVPEPPSDADQLQPKVSICGLD----------------MP
MAV_3758|M.avium_104 ----MTDTADSS-NPQSHAGDADPKVSTRNPDTRPD--------DDAGAP
MAB_3223c|M.abscessus_ATCC_199 DETGLTAWDPQAEKVFGDVTLDDSRQSLGQFETVGDNVEHDAAEKVSAVD
:* :.
Mflv_4169|M.gilvum_PYR-GCK DPVPAEEPADEE--KTGKK-RGALREAAILISIALVLYYVTLTFIARPYL
Mvan_2193|M.vanbaalenii_PYR-1 DPESPEPLAAEGDGKTGKKKRGALREAAILISIALVLYYVMLTFIARPYL
MSMEG_2441|M.smegmatis_MC2_155 PADEPQSRPDPEQDEKPKRKHGALREGAILVTIAVVLYYVMLTFIARPYL
Mb2927c|M.bovis_AF2122/97 DADSEGDSKAAKTDEPRPAKRSTLREFAVLAVIAVVLYYVMLTFVARPYL
Rv2903c|M.tuberculosis_H37Rv DADSEGDSKAAKTDEPRPAKRSTLREFAVLAVIAVVLYYVMLTFVARPYL
MMAR_1805|M.marinum_M QEEPGAESRAADEDESKPAKKSTLRELATLAVIAIVIYYVMLTFVARPYL
MUL_2053|M.ulcerans_Agy99 QEELGAESRAADEDESKPAKKSTLRELATLAVIAIVIYYVMLTFVARPYL
MLBr_01612|M.leprae_Br4923 AEVSET-AAEAAIGVSEPKKRSALWEFAILAVIAIGLYYVMLTFVARPYL
MAV_3758|M.avium_104 APVPEPGTKPDAAAPTEGSKRSTLREFAILAVIAVLLYYVMLTFVARPYL
MAB_3223c|M.abscessus_ATCC_199 STDEDPAQDSKDASDSDEGGSGMLREVGVLLLIALVLYCITQNFIARPYL
. * * . * **: :* : .*:*****
Mflv_4169|M.gilvum_PYR-GCK IPSESMEPTLHGCNGCVGDRIMVDKMSYRFGSPEPGDVVVFKGPPNWSIG
Mvan_2193|M.vanbaalenii_PYR-1 IPSESMEPTLHGCNGCVGDRIMVDKLTYRFSSPEPGDVVVFKGPPNWSVG
MSMEG_2441|M.smegmatis_MC2_155 IPSESMEPTLHGCNGCVGDRIMVDKLTYRFTEPRPGDVVVFKGPPSWNIG
Mb2927c|M.bovis_AF2122/97 IPSESMEPTLHGCSTCVGDRIMVDKLSYRFGSPQPGDVIVFRGPPSWNVG
Rv2903c|M.tuberculosis_H37Rv IPSESMEPTLHGCSTCVGDRIMVDKLSYRFGSPQPGDVIVFRGPPSWNVG
MMAR_1805|M.marinum_M IPSESMEPTLHGCSTCVGDRIMVDKLTYRFTSPKPGDVIVFKGPPSWNVG
MUL_2053|M.ulcerans_Agy99 IPSESMEPTLHGCSTCVGDRIMVDKLTYRFTSPKPGDVIVFKGPPSWNVG
MLBr_01612|M.leprae_Br4923 IPSESMEPTLHGCSGCVGDRIMVDKITYRFSSPQPGDVIVFKGPPSWNTM
MAV_3758|M.avium_104 IPSESMEPTLHGCTGCVGDRIMVDKVSYRFGAPRPGDVIVFKGPPSWNLG
MAB_3223c|M.abscessus_ATCC_199 IPSESMEPTLHGCRGCTGDRIMVDKVVFRFSEPQPGEVVVFKGPPEWNGN
************* *.********: :** *.**:*:**:***.*.
Mflv_4169|M.gilvum_PYR-GCK YKSIRSDNAAVRWIQDTLSVVGFVPPDQNDLVKRIIATGGQTVQCRVDTG
Mvan_2193|M.vanbaalenii_PYR-1 YKSIRSDNTAVRWVQNALSVVGFVPPDENDLVKRIIAVGGQTVQCRVDTG
MSMEG_2441|M.smegmatis_MC2_155 YKSIRSDNPVIRGVQNALSFIGFVPPDENDLVKRVIAVGGQTVECRAATG
Mb2927c|M.bovis_AF2122/97 YKSIRSHNVAVRWVQNALSFIGFVPPDENDLVKRVIAVGGQTVQCRSDTG
Rv2903c|M.tuberculosis_H37Rv YKSIRSHNVAVRWVQNALSFIGFVPPDENDLVKRVIAVGGQTVQCRSDTG
MMAR_1805|M.marinum_M YKSIRSSNTALRWVQNALSFIGFVPPDENDLVKRVIAVGGQTVQCRSDTG
MUL_2053|M.ulcerans_Agy99 YKSIRSSNTALRWVQNALSFIGFVPPDENDLVKRVIAVGGQTVQCRSDTG
MLBr_01612|M.leprae_Br4923 YKSIRSNNIVLRSVQNALSFVGFVPPDENDLVKRVIAVGGQTVQCRSDTG
MAV_3758|M.avium_104 YKSIRSNNTALRWAQNALSFVGFVPPDENDLVKRVIAVGGQTVQCRAETG
MAB_3223c|M.abscessus_ATCC_199 YRSIRSSNTAVRYVQNALSFIGVVPPDENDLVKRVIATGGQTVECRPNTG
*:**** * .:* *::**.:*.****:******:**.*****:** **
Mflv_4169|M.gilvum_PYR-GCK LTVDGKPLNEPYLNAETMMADPAVYPCLGNEFGPVTVPEGRLWVMGDNRT
Mvan_2193|M.vanbaalenii_PYR-1 LTVDGKPLNEPYLDPDTMMADPAVYPCLGNEFGPVTVPEGRLWVMGDNRT
MSMEG_2441|M.smegmatis_MC2_155 LTVDGKKLDEPYLDPTTMMADPAIYPCLGNEFGPVTVPEDRIWVMGDNRT
Mb2927c|M.bovis_AF2122/97 LTVNGRPLKEPYLDPATMMADPSIYPCLGSEFGPVTVPPGRVWVMGDNRT
Rv2903c|M.tuberculosis_H37Rv LTVNGRPLKEPYLDPATMMADPSIYPCLGSEFGPVTVPPGRVWVMGDNRT
MMAR_1805|M.marinum_M LTVDGKPLKEPYLDPATMLADPSVYPCLGSEFGPVTVPAGRLWVMGDNRT
MUL_2053|M.ulcerans_Agy99 LTVDGKPLKEPYLDPATMLADPSVYPCLGSEFGPVTVPAGRLWVMGDNRT
MLBr_01612|M.leprae_Br4923 LTVNGKPLKEPYLRPVTMNADLSFSPCLGSEFGPVTVPQGRLWVMGDNRI
MAV_3758|M.avium_104 LTVNGKPLREPYLDRNTMAADPSVYPCLGSEFGPVTVPAGRLWVMGDNRT
MAB_3223c|M.abscessus_ATCC_199 LTVNGKKLDEPYLDPETIG--PNIDGCWGFPFGPVKVPEGKLWMMGDNRT
***:*: * **** *: . * * ****.** .::*:*****
Mflv_4169|M.gilvum_PYR-GCK HSADSRTHCTNEPADV------QKGLLCTGDPTAGTIPVENVIGKAQFIA
Mvan_2193|M.vanbaalenii_PYR-1 HSADSRTHCSNVPADA------QRGLLCTGDPAAGTIPEENVIGKARFIA
MSMEG_2441|M.smegmatis_MC2_155 HSADSRVHCTNLPADA------QKGLLCTGDPTAGTIPVENVIGKARFIA
Mb2927c|M.bovis_AF2122/97 HSADSRAHCPLL---------------CTNDPLPGTVPVANVIGKARLIV
Rv2903c|M.tuberculosis_H37Rv HSADSRAHCPLL---------------CTDDPLPGTVPVANVIGKARLIV
MMAR_1805|M.marinum_M HSADSRAHCPML---------------CTGDAMAGTVPVANVIGKARFIV
MUL_2053|M.ulcerans_Agy99 HSADSRAHCPML---------------CTGDAMAGTVPVANVIGKARFIV
MLBr_01612|M.leprae_Br4923 HSADSRYHCNST--DV------VNGLSCTGDPNSGTVPVSNVIGKARVVV
MAV_3758|M.avium_104 HSADSRAHCTSVPAEA------LKGVLCTGDPASGTVPVSNVIGKARFIV
MAB_3223c|M.abscessus_ATCC_199 HSGDSRAHCQSRDRDAGLDVDIDHKIYCTGDPNIGTVPVANVIGKARFIA
**.*** ** **.*. **:* ******:.:.
Mflv_4169|M.gilvum_PYR-GCK WPPGRWGGVNSVNPQS--
Mvan_2193|M.vanbaalenii_PYR-1 WPPGRWGGVSSVNPQS--
MSMEG_2441|M.smegmatis_MC2_155 WPPSRWGGISDVNPQTSS
Mb2927c|M.bovis_AF2122/97 WPPSRWGVVRSVNPQQGR
Rv2903c|M.tuberculosis_H37Rv WPPSRWGVVRSVNPQQGR
MMAR_1805|M.marinum_M WPPSRWGVVRSVNPQTGQ
MUL_2053|M.ulcerans_Agy99 WPPSRWGVVRSVNPQTGQ
MLBr_01612|M.leprae_Br4923 WPPSRWGGVGSVNSQQGQ
MAV_3758|M.avium_104 WPPSRWGGVGSVNPQQGQ
MAB_3223c|M.abscessus_ATCC_199 WPPGRWGAVKTVNPQT--
***.*** : **.*