For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRRTLYVQLIIAFMCVAMVAYLVMLGRVAVAMIGSGRAAAAGLGLALLILPVIGLWAMIATLRAGFAYQ RLARLIAEDGLDIDASALPRRASGRIQRDAADALFAAVRTELEDDADDWRRWYRLARAYDYAGDRRRARE AMKTALQLEGRARPGAR
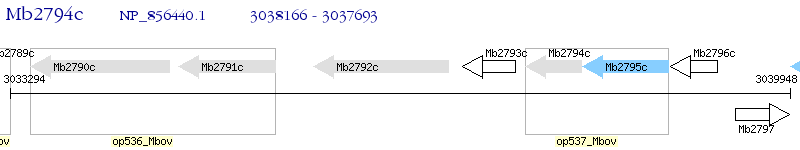
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2794c | - | - | 100% (157) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4012 | - | 1e-55 | 66.67% (150) | TPR repeat-containing protein |
| M. tuberculosis H37Rv | Rv2772c | - | 2e-84 | 100.00% (157) | transmembrane protein |
| M. leprae Br4923 | MLBr_01526 | - | 8e-63 | 76.62% (154) | hypothetical protein MLBr_01526 |
| M. abscessus ATCC 19977 | MAB_3095c | - | 9e-49 | 61.27% (142) | hypothetical protein MAB_3095c |
| M. marinum M | MMAR_1939 | - | 3e-64 | 77.33% (150) | hypothetical protein MMAR_1939 |
| M. avium 104 | MAV_3661 | - | 2e-62 | 74.83% (147) | TPR repeat-containing protein |
| M. smegmatis MC2 155 | MSMEG_2665 | - | 6e-53 | 66.44% (149) | hypothetical protein MSMEG_2665 |
| M. thermoresistible (build 8) | TH_0696 | - | 3e-55 | 67.33% (150) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2165 | - | 9e-64 | 76.00% (150) | hypothetical protein MUL_2165 |
| M. vanbaalenii PYR-1 | Mvan_2375 | - | 2e-54 | 67.33% (150) | tetratricopeptide TPR_2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4012|M.gilvum_PYR-GCK MSD------GSRALRIQLLIGFMCVALVVYFVLLGRTALILIGSGEAAPV
Mvan_2375|M.vanbaalenii_PYR-1 MSE------GARALRIQLLIGFMCVALVVYFVLLGRAALMLITSGEPAPI
MSMEG_2665|M.smegmatis_MC2_155 MSD------GARSLRIQLLIGFMCVALIVYFVMLGRAAFVFITSGEAAAV
TH_0696|M.thermoresistible__bu MSDPDSNDGGGRALRIQLLIGFMCVALVVYFVLLGRLGWMFITSGSAAAV
Mb2794c|M.bovis_AF2122/97 --MTRRT------LYVQLIIAFMCVAMVAYLVMLGRVAVAMIGSGRAAAA
Rv2772c|M.tuberculosis_H37Rv --MTRRT------LYVQLIIAFMCVAMVAYLVMLGRVAVAMIGSGRAAAA
MMAR_1939|M.marinum_M MSDTVRT------VRVQVLIAFMCVALVAYFVMLGRLAFAMIGSGRAAAV
MUL_2165|M.ulcerans_Agy99 MSDTVRT------VRVQVLIAFMCVALLAYFVMLGRLAFAMIGSGRAAAV
MLBr_01526|M.leprae_Br4923 MNDTVMTKRSLYILYIQLLIAALCFAMLAYLVLLGRMAVAMIGSGQAAAV
MAV_3661|M.avium_104 MNKA---------LRIQLLVAFLCVALLVYFALLGRVAIAMIASGRAAAV
MAB_3095c|M.abscessus_ATCC_199 MSAA---------ARVKILIAFMCVAMVVYFLILGKMGIALVQTGKAAAI
:::::. :*.*::.*: :**: . :: :* .*.
Mflv_4012|M.gilvum_PYR-GCK GLGLAILIMPAIGAWVLVATLKAGLAHQRLARKARESGMDLDVDDLPRRA
Mvan_2375|M.vanbaalenii_PYR-1 GLGLAILILPVIGVWVMVATLKAGLAHQRLARLAREQGMELDVSGLPRRP
MSMEG_2665|M.smegmatis_MC2_155 ALGIALLAFPVIGVWVLIATLRAGLAHQRLGRLADEQGMDLDVSDLPRRP
TH_0696|M.thermoresistible__bu GLGLALMILPFIGVWAMVSTLRDGLAHQRLARIIGEEGFELDVSDLPRRP
Mb2794c|M.bovis_AF2122/97 GLGLALLILPVIGLWAMIATLRAGFAYQRLARLIAEDGLDIDASALPRRA
Rv2772c|M.tuberculosis_H37Rv GLGLALLILPVIGLWAMIATLRAGFAYQRLARLIAEDGLDIDASALPRRA
MMAR_1939|M.marinum_M GLGLALLIMPLIGLWAMVATLRAGFTHQKLARLIAEEGLELDADGLPRRA
MUL_2165|M.ulcerans_Agy99 GLGLALLIMPLIGLWAMVATLRAGFTHQKLARLIAEEGLELDADGLPRRA
MLBr_01526|M.leprae_Br4923 SLGLALLIMPVIGLWAMIATLRAGFAHQKLARLIAADGMELDTSVLLRRP
MAV_3661|M.avium_104 GLGVAVLIMPALGLWAMIATLRAGFAHQRLARLIAADGMELDTSALPRRP
MAB_3095c|M.abscessus_ATCC_199 GMGVGVLILPFIGLWAMIATLRAGFAHQRLAALAAQQGRELDVSALRTTP
.:*:.:: :* :* *.:::**: *:::*:*. .* ::*.. * .
Mflv_4012|M.gilvum_PYR-GCK SGRIERDAADALFATVRTEVENDPDNWVRWYRLARAYDYAGDRTRARETM
Mvan_2375|M.vanbaalenii_PYR-1 SGRIERDAADALFATVRDELEADPDNWLRWYRLARAYDYAGDRGRARETM
MSMEG_2665|M.smegmatis_MC2_155 SGRIERDAADALFVQVKAELEADPDNWVRWYRLARAYDYAGDRSRARDAM
TH_0696|M.thermoresistible__bu SGRIQREAADALFEQVRAELEADPDNWRRWYRVARAYDYAGDRRRAREAM
Mb2794c|M.bovis_AF2122/97 SGRIQRDAADALFAAVRTELEDDADDWRRWYRLARAYDYAGDRRRAREAM
Rv2772c|M.tuberculosis_H37Rv SGRIQRDAADALFAAVRTELEDDADDWRRWYRLARAYDYAGDRRRAREAM
MMAR_1939|M.marinum_M SGRIERDAADALFDTMRAELEEDPQNWRRWYRLARAYDYAGDRRRAREAM
MUL_2165|M.ulcerans_Agy99 SGRIERDAADALFDTMRAELEEDPQNWRRWYRLARAYDYAGDRRRAREAM
MLBr_01526|M.leprae_Br4923 SGRIQRDAADALFATVRAELAGDPDNWRCWYRLARAYDYAGDRRRAREAM
MAV_3661|M.avium_104 SGRIRRDAADALFATVRTEVENHPDDWRRWYRLARAYDYAGDRRRAREAM
MAB_3095c|M.abscessus_ATCC_199 SGRVNREDADALFATVKAEVEAEPDDWVGWYRLARAYDYAGDRGRARDTM
***:.*: ***** :: *: ..::* ***:********** ***::*
Mflv_4012|M.gilvum_PYR-GCK KKAVQMEESAQ----
Mvan_2375|M.vanbaalenii_PYR-1 KKAVLLQEAAR----
MSMEG_2665|M.smegmatis_MC2_155 RTAVRMQEGRR----
TH_0696|M.thermoresistible__bu RKAVELQERDR----
Mb2794c|M.bovis_AF2122/97 KTALQLEGRARPGAR
Rv2772c|M.tuberculosis_H37Rv KTALQLEGRARPGAR
MMAR_1939|M.marinum_M KTAVELQARQ-----
MUL_2165|M.ulcerans_Agy99 RTAVELQARQ-----
MLBr_01526|M.leprae_Br4923 KTALELHGRD-----
MAV_3661|M.avium_104 KTAVDLQGHQ-----
MAB_3095c|M.abscessus_ATCC_199 RTAVQMQQDAAR---
:.*: :.