For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNAYDVLKRHHTVLKGLGRKVGEAPVNSEERHVLFDEMLIELDIHFRIEDDLYYPALSAAGKPITGTHAE HRQVVDQLATLLRTPQRAPGYEEEWNVFRTVLEAHADVEERDMIPAPTPVHITDAELEELGDKMAARIEQ LRGSPLYTLRTKGKADLLKAI
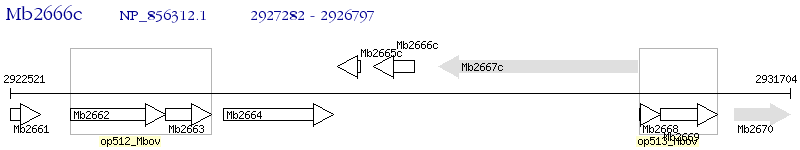
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2666c | - | - | 100% (161) | hypothetical protein Mb2666c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2633c | - | 2e-90 | 100.00% (161) | hypothetical protein Rv2633c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3507 | - | 3e-69 | 75.78% (161) | hemerythrin HHE cation binding domain-containing protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2666c|M.bovis_AF2122/97 MNAYDVLKRHHTVLKGLGRKVGEAPVNSEERHVLFDEMLIELDIHFRIED
Rv2633c|M.tuberculosis_H37Rv MNAYDVLKRHHTVLKGLGRKVGEAPVNSEERHVLFDEMLIELDIHFRIED
MAV_3507|M.avium_104 MNAYDVLKEHHIVIKGLGRKISEAPVNSEERHALFDDLLIELDIHFRIED
********.** *:******:.**********.***::************
Mb2666c|M.bovis_AF2122/97 DLYYPALSAAGKPITGTHAEHRQVVDQLATLLRTPQRAPGYEEEWNVFRT
Rv2633c|M.tuberculosis_H37Rv DLYYPALSAAGKPITGTHAEHRQVVDQLATLLRTPQRAPGYEEEWNVFRT
MAV_3507|M.avium_104 DLYYPALKEASKLIAVAHAEHRQVVDQLSVLLRTPQSAPNYEDEWNSFKT
*******. *.* *: :***********:.****** **.**:*** *:*
Mb2666c|M.bovis_AF2122/97 VLEAHADVEERDMIPAPTPVHITDAELEELGDKMAARIEQLRGSPLYTLR
Rv2633c|M.tuberculosis_H37Rv VLEAHADVEERDMIPAPTPVHITDAELEELGDKMAARIEQLRGSPLYTLR
MAV_3507|M.avium_104 VLEAHADEEERDMIPAPPQVHITDAELEELGNKMAAKMEQYRGSALYKMR
******* *********. ************:****::** ***.**.:*
Mb2666c|M.bovis_AF2122/97 TKGKADLLKAI
Rv2633c|M.tuberculosis_H37Rv TKGKADLLKAI
MAV_3507|M.avium_104 TKGRAALVRSL
***:* *::::