For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDRSREPADPWKGFSAVMAATLILEAIVVLLAIPVVDAVGGGLRPASLGYLVGLAVLLILLTGLQRRPW AIWVNLGAQPVLVAGFAVYPGVGFIGVLFAALWVLIAYLRAEVRRRRDYRVSQ
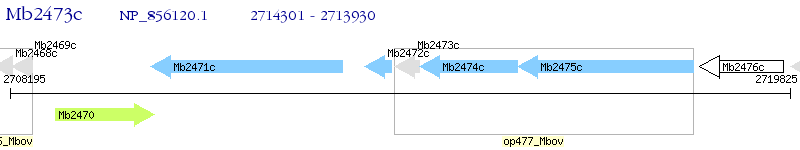
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2473c | - | - | 100% (123) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2632 | - | 8e-39 | 65.49% (113) | hypothetical protein Mflv_2632 |
| M. tuberculosis H37Rv | Rv2446c | - | 5e-66 | 100.00% (123) | integral membrane protein |
| M. leprae Br4923 | MLBr_01470 | - | 3e-36 | 66.67% (108) | hypothetical protein MLBr_01470 |
| M. abscessus ATCC 19977 | MAB_1605 | - | 1e-34 | 61.82% (110) | hypothetical protein MAB_1605 |
| M. marinum M | MMAR_3770 | - | 4e-44 | 68.80% (125) | hypothetical protein MMAR_3770 |
| M. avium 104 | MAV_1726 | - | 2e-48 | 79.82% (114) | hypothetical protein MAV_1726 |
| M. smegmatis MC2 155 | MSMEG_4628 | - | 5e-34 | 60.87% (115) | hypothetical protein MSMEG_4628 |
| M. thermoresistible (build 8) | TH_3032 | - | 8e-31 | 50.00% (116) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3717 | - | 3e-43 | 67.20% (125) | hypothetical protein MUL_3717 |
| M. vanbaalenii PYR-1 | Mvan_3949 | - | 6e-41 | 69.09% (110) | hypothetical protein Mvan_3949 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2632|M.gilvum_PYR-GCK ---------------MS-TPPPDPWKSFRGVMAGTLILEAIVVLLALPVV
Mvan_3949|M.vanbaalenii_PYR-1 ---------------MNGQPPPDPWKSFRGVMAGTLILEAIVVLLALPVV
MAB_1605|M.abscessus_ATCC_1997 MSDPEDGIEPRADQNVAGAAPPDPWRSFRGVMAGTLILETIVVLLAIPVV
Mb2473c|M.bovis_AF2122/97 MTD----------RSREPA---DPWKGFSAVMAATLILEAIVVLLAIPVV
Rv2446c|M.tuberculosis_H37Rv MTD----------RSREPA---DPWKGFSAVMAATLILEAIVVLLAIPVV
MAV_1726|M.avium_104 MTRPDPPEGP---QQRGAA---DPWRSFGAVMAATLILEAIVVLLAIPVV
MMAR_3770|M.marinum_M MTG-QPQEPA---NHGDPAPQSDPWKSFGSVMAATLILEAIVVLLAIPVV
MUL_3717|M.ulcerans_Agy99 MTG-QPQEPA---NHGDPAPQSDPWKSFGSVMTATLTLEAIVVLLAIPVV
MLBr_01470|M.leprae_Br4923 MTDSPGEHGP---QKPLPS--ADPWKSFAGVMAAILFLEAIVVLLALPVL
MSMEG_4628|M.smegmatis_MC2_155 MSENQA-------PDNEPAPAKDPWKSFRGVMAACLILEAIVVLLALPVV
TH_3032|M.thermoresistible__bu MSE-------------QPARRPDPWRSFRGVMAGTLLLEAIVMVLALPVV
***:.* .**:. * **:**::**:**:
Mflv_2632|M.gilvum_PYR-GCK GAVGPGLTPFSTAYVIGFAVLLVVMAGVQGRGWAIWANVGVQLLLVAGWF
Mvan_3949|M.vanbaalenii_PYR-1 GAVGPGLTAGSTGYVIGVAVLLVLMSGVQGRPWAIWANLGVQLLLVAGWF
MAB_1605|M.abscessus_ATCC_1997 KMVGGGLSPLSLSYLIGVVVALVLLCGMQGRPWALQANLALQLVLVAGFL
Mb2473c|M.bovis_AF2122/97 DAVGGGLRPASLGYLVGLAVLLILLTGLQRRPWAIWVNLGAQPVLVAGFA
Rv2446c|M.tuberculosis_H37Rv DAVGGGLRPASLGYLVGLAVLLILLTGLQRRPWAIWVNLGAQPVLVAGFA
MAV_1726|M.avium_104 GAVGGGLTRVSLGYLIGLAVLLVLLAGVQRRPWAIWVNLGAQAILLAGFA
MMAR_3770|M.marinum_M GAVGGGLTPASLGYLIGLAVTLVLLAGVQRRPWAIWANLGIQLVPIAGVF
MUL_3717|M.ulcerans_Agy99 GAVGGGLTPASLGYLIGLAVTLVLLAGVQRRPWAIWANLGIQLVPIAGVF
MLBr_01470|M.leprae_Br4923 GASGG-LTLSALSFLIGLAGLLIVLVGLQRKAWAIWVNLGVQVVVLVGCA
MSMEG_4628|M.smegmatis_MC2_155 AVG--GLSAAAGGYLIGLAVVLILMCGVQGRPWALWANLGMQVPVIAGAV
TH_3032|M.thermoresistible__bu VQAQNGFTATGGGFVVALTLALVALAGVQGRPWALWVNIAMQFVVIAGVL
: . .:::... *: : *:* : **: .*:. * :.*
Mflv_2632|M.gilvum_PYR-GCK LYPGVGFIGLLFFVVWLLIAYLRAEVLRRQKRGLLPGQSG--GDH
Mvan_3949|M.vanbaalenii_PYR-1 IYPGVGFIGLLFVVVWLLIAYLRAEVLRRQKRGLLPGQQP--TDD
MAB_1605|M.abscessus_ATCC_1997 VNAAVGFVGLLFLGVWLLIWYMRNQVRDRQSRGELPGQQGSATDQ
Mb2473c|M.bovis_AF2122/97 VYPGVGFIGVLFAALWVLIAYLRAEVRRR--RDYRVSQ-------
Rv2446c|M.tuberculosis_H37Rv VYPGVGFIGVLFAALWVLIAYLRAEVRRR--RDYRVSQ-------
MAV_1726|M.avium_104 VYPGVGFIGVLFTGLWALIAYFRAEVRRR--QG------------
MMAR_3770|M.marinum_M VYPAVGFLGVVFGAVWAMMAYFRAEVRRRERQGPPPSR-------
MUL_3717|M.ulcerans_Agy99 VYPAVGFLGVVFGAVWAMMAYFRAEVRRRERQGPPPSR-------
MLBr_01470|M.leprae_Br4923 VYPVLGFVGVLFAGLWALIVYFRAEVSRR----------------
MSMEG_4628|M.smegmatis_MC2_155 FHGGIGFIGVVFLVVWLLIVYLRHEVKRRQERGLLPGQQRPAE--
TH_3032|M.thermoresistible__bu VHGAIGFVGVLFLAVWLFIAYLRAEVKRREARGELPGQQAGEE--
. :**:*::* :* :: *:* :* *