For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSNNAGESDGTGPALPTVWPGNAYPLGATYDGAGTNFSLFSEIAEKVELCLIDEDGVESRIPLDEVDGY VWHAYLPNITPGQRYGFRVHGPFDPAAGHRCDPSKLLLDPYGKSFHGDFTFGQALYSYDVNAVDPDSTPP MVDSLGHTMTSVVINPFFDWAYDRSPRTPYHETVIYEAHVKGMTQTHPSIPPELRGTYAGLAHPVIIDHL NELNVTAVELMPVHQFLHDSRLLDLGLRNYWGYNTFGFFAPHHQYASTRQAGSAVAEFKTMVRSLHEAGI EVILDVVYNHTAEGNHLGPTINFRGIDNTAYYRLMDHDLRFYKDFTGTGNSLNARHPHTLQLIMDSLRYW VIEMHVDGFRFDLASTLARELHDVDRLSAFFDLVQQDPVVSQVKLIAEPWDVGEGGYQVGNFPGLWTEWN GKYRDTVRDYWRGEPATLGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLNDLVSYNDKHNEANGE NNRDGESYNRSWNCGVEGPTDDPDILALRARQMRNMWATLMVSQGTPMIAHGDEIGRTQYGNNNVYCQDS ELSWMDWSLVDKNADLLAFARKATTLRKNHKVFRRRRFFEGEPIRSGDEVRDIAWLTPSGREMTHEDWGR GFDRCVAVFLNGEAITAPDARGERVVDDSFLLCFNAHDHDVEFVMPHDGYAQQWTGELDTNDPVGDIDLT VTATDTFSVPARSLLVLRKTL
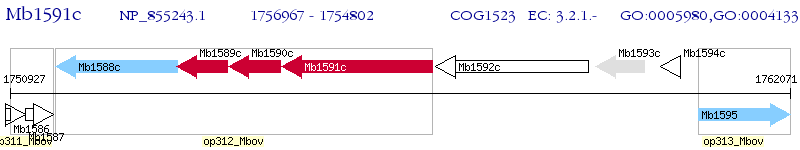
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1591c | treX | - | 100% (721) | maltooligosyltrehalose synthase TreX |
| M. bovis AF2122 / 97 | Mb1361c | glgB | 2e-11 | 26.15% (348) | glycogen branching enzyme |
| M. bovis AF2122 / 97 | Mb1588c | treZ | 7e-09 | 25.63% (277) | maltooligosyltrehalose trehalohydrolase TreZ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3631 | - | 0.0 | 82.34% (708) | glycogen debranching enzyme GlgX |
| M. tuberculosis H37Rv | Rv1564c | treX | 0.0 | 100.00% (721) | maltooligosyltrehalose synthase TreX |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2690 | - | 0.0 | 77.62% (706) | glycogen operon protein GlgX-like protein |
| M. marinum M | MMAR_2379 | treX | 0.0 | 86.46% (724) | maltooligosyltrehalose synthase TreX |
| M. avium 104 | MAV_3210 | glgX | 0.0 | 83.27% (735) | glycogen debranching enzyme GlgX |
| M. smegmatis MC2 155 | MSMEG_3186 | glgX | 0.0 | 82.79% (703) | glycogen debranching enzyme GlgX |
| M. thermoresistible (build 8) | TH_1798 | glgX | 0.0 | 74.58% (708) | Probable Maltooligosyltrehalose synthase TreX |
| M. ulcerans Agy99 | MUL_1555 | treX | 0.0 | 86.19% (724) | maltooligosyltrehalose synthase TreX |
| M. vanbaalenii PYR-1 | Mvan_2786 | - | 0.0 | 80.70% (710) | glycogen debranching enzyme GlgX |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3631|M.gilvum_PYR-GCK MSS----------AQPASVPTVWPGTPYPLGATYDGAGTNFSVFSEVAER
Mvan_2786|M.vanbaalenii_PYR-1 MSS----------AGPS-APTVWPGTPYPLGATYDGAGTNFSVFSEVAER
MSMEG_3186|M.smegmatis_MC2_155 MSDP-------TASPPAPMNMVWPGEAYPLGATYDGAGTNFSLFSEVAER
Mb1591c|M.bovis_AF2122/97 MSSN---NAGESDGTGPALPTVWPGNAYPLGATYDGAGTNFSLFSEIAEK
Rv1564c|M.tuberculosis_H37Rv MSSN---NAGESDGTGPALPTVWPGNAYPLGATYDGAGTNFSLFSEIAEK
MMAR_2379|M.marinum_M MSPN---NAVDSGVTPPPLPTVWPGSAYPLGATYDGAGTNFSLFSEIADR
MUL_1555|M.ulcerans_Agy99 MSPN---NAVDSGVTPPPLPTVWPGSAYPLGATYDGAGTNFSLFSEIADR
MAV_3210|M.avium_104 MSSNEPASAAGGGTHQPEVPTVWPGSPYPLGASYDGAGTNFSLFSEIAEK
MAB_2690|M.abscessus_ATCC_1997 -------------MTLPPL-QVWPGNPYPLGATYDGAGTNFSLFSEVATS
TH_1798|M.thermoresistible__bu ----------VTSTAPTSVPMVWPGSAYPLGATYDGAGTNFSVFSEVADR
. **** .*****:*********:***:*
Mflv_3631|M.gilvum_PYR-GCK VELCLIGKDGREERIDLDEVDGFVWHCYLPTITPGQRYGFRVHGPWDPAA
Mvan_2786|M.vanbaalenii_PYR-1 VELCLVGKDGREQRIDLDEVDGFVWHCYLPTVTPGQRYGFRVHGPWDPGA
MSMEG_3186|M.smegmatis_MC2_155 VELCLIAKDGTETRINLDEIDGYVWHAYLPTISPGQRYGFRVYGPWDPSK
Mb1591c|M.bovis_AF2122/97 VELCLIDEDGVESRIPLDEVDGYVWHAYLPNITPGQRYGFRVHGPFDPAA
Rv1564c|M.tuberculosis_H37Rv VELCLIDEDGVESRIPLDEVDGYVWHAYLPNITPGQRYGFRVHGPFDPAA
MMAR_2379|M.marinum_M VELCLIADDDSESRISLEEVDGYVWHAYLPNISPGQRYGFRVYGPFDPAA
MUL_1555|M.ulcerans_Agy99 VELCLIADDDSESRISLEEVDGYVWHAYLPNISPGQRYGFRVYGPFDPAA
MAV_3210|M.avium_104 VELCLIDSRGAETRIPLDEVDGYVWHAYLPNINPGQRYGFRVYGPFEPSA
MAB_2690|M.abscessus_ATCC_1997 VELCLIAKDGTETRIPLDEVDGYVWHCYLPTISPGQRYGFRVHGPWDPES
TH_1798|M.thermoresistible__bu VELCLIARDGAETRINLEEVDGYVWHCYLPAVTPGQRYGYRVHGPWNPAT
*****: . * ** *:*:**:***.*** :.******:**:**::*
Mflv_3631|M.gilvum_PYR-GCK GHRCDASKLLLDPYGKSFHGDFEFG----QALYSYDLEADDL----ATGG
Mvan_2786|M.vanbaalenii_PYR-1 GHRCDPSKLLLDPYGKSFHGDFDFS----QALYSYDLRADDL----ASGG
MSMEG_3186|M.smegmatis_MC2_155 GLRCDPSKLLLDPYGKSFHGDFDFS----QALYSYDLEADPP----GSG-
Mb1591c|M.bovis_AF2122/97 GHRCDPSKLLLDPYGKSFHGDFTFG----QALYSYDVNAVDP--DST---
Rv1564c|M.tuberculosis_H37Rv GHRCDPSKLLLDPYGKSFHGDFTFG----QALYSYDVNAVDP--DST---
MMAR_2379|M.marinum_M GHRCDPSKLLLDPYGKAFDGDFTFG----QALFSYDMKTVDL--GSADPG
MUL_1555|M.ulcerans_Agy99 GHRCDPSKLLLDPYGKAFDGDFTFG----QALFSYDMKTVDL--GSADPG
MAV_3210|M.avium_104 GHRCDPSKLLLDPYGKAFHGDFTYG----QALFSYDLKAVAAGGDDADPG
MAB_2690|M.abscessus_ATCC_1997 GHRCDPSKLLLDPYGKAFHGEFGYVPDTAPPLLSYQIDPVDT-------E
TH_1798|M.thermoresistible__bu GQRCDPSKLLLDPYGKTFDGAFRWG----QALFSFDLEAEDP----ASGG
* ***.**********:*.* * : .* *::: .
Mflv_3631|M.gilvum_PYR-GCK VPPRVDSLGHTMTSVVINPFFQWGSDRAPRTSYHETVIYEAHVKGMTQTH
Mvan_2786|M.vanbaalenii_PYR-1 VPPQVDSLGHTMTSVVINPFFQWGSDRPPRTPYHETVIYEAHVKGMTQTH
MSMEG_3186|M.smegmatis_MC2_155 TPPRVDSLGHTMTSVVINPFFQWGSDRAPKTPYHDTVIYEAHVKGMTQTH
Mb1591c|M.bovis_AF2122/97 -PPMVDSLGHTMTSVVINPFFDWAYDRSPRTPYHETVIYEAHVKGMTQTH
Rv1564c|M.tuberculosis_H37Rv -PPMVDSLGHTMTSVVINPFFDWAYDRSPRTPYHETVIYEAHVKGMTQTH
MMAR_2379|M.marinum_M IPPMVDSLGSTMTSVVINPFFDWGYDRAPMIPYHETVIYEAHVKGMTQTH
MUL_1555|M.ulcerans_Agy99 IPPMVDSLGSTMTSVVINPFFDWGYDRAPMIPYHETVIYEAHVKGMTQTH
MAV_3210|M.avium_104 IPPMVDSLGHTMTSVVSNPFFDWGSDRAPLTPYHETVIYEAHVKGMTQTH
MAB_2690|M.abscessus_ATCC_1997 TLVPRDSLGRTMTTVVINPYFDWGSDRRPRTPYHETVIYEAHVKGMTQTH
TH_1798|M.thermoresistible__bu NPPMVDSLGHTMTSVVTNPFFDWGSDRPPKIPYHETIIYEAHVKGMTQTH
**** ***:** **:*:*. ** * .**:*:*************
Mflv_3631|M.gilvum_PYR-GCK PGVPEELRGTYVGLGHPVIIDHLKSLNVTAIELMPVHQFMHDHRLLDLGL
Mvan_2786|M.vanbaalenii_PYR-1 PGIPEELRGTYAGLGHPAIIDHLKSLNVTAIELMPVHQFLHDHRLLDLGL
MSMEG_3186|M.smegmatis_MC2_155 PGVPEALRGTYAGLSHPVIIEHLQSLNVTAIELMPVHQFMHDHRLLDLGL
Mb1591c|M.bovis_AF2122/97 PSIPPELRGTYAGLAHPVIIDHLNELNVTAVELMPVHQFLHDSRLLDLGL
Rv1564c|M.tuberculosis_H37Rv PSIPPELRGTYAGLAHPVIIDHLNELNVTAVELMPVHQFLHDSRLLDLGL
MMAR_2379|M.marinum_M PDIPEELRGTYAGLAHPAIIDHLKSLNVTTIELMPVHQFMHDSRLLDLGL
MUL_1555|M.ulcerans_Agy99 PDIPEELRGTYAGLAHPAIIDHLKSLNVTTIELMPVHQFMHDSRLLDLGL
MAV_3210|M.avium_104 PSVPEQLRGTYAGLAHPAIIDHLKSLNVTAIELMPVHQFMHDSRLLDLGL
MAB_2690|M.abscessus_ATCC_1997 PGIPEELRGTYAGLAHPAVIDHLHSLGVTAIELMPVHQFFHDSRLIALGL
TH_1798|M.thermoresistible__bu PGVPEPLRGTYAGLCHPAIIEHLQSLHVTAIELMPVHQFMHDQRLLDLGL
*.:* *****.** **.:*:**:.* **::********:** **: ***
Mflv_3631|M.gilvum_PYR-GCK RNYWGYNTFGFFAPHNQYAATRSAGGAVAEFKTMVRAFHEAGIEVILDVV
Mvan_2786|M.vanbaalenii_PYR-1 RNYWGYNTFGFFAPHYQYAANRNAGGAVGEFKTMVREFHEAGIEVILDVV
MSMEG_3186|M.smegmatis_MC2_155 RNYWGYNTVGFFAPHFQYAATRNAGGAVAEFKTMVKAFHDAGIEVILDVV
Mb1591c|M.bovis_AF2122/97 RNYWGYNTFGFFAPHHQYASTRQAGSAVAEFKTMVRSLHEAGIEVILDVV
Rv1564c|M.tuberculosis_H37Rv RNYWGYNTFGFFAPHHQYASTRQAGSAVAEFKTMVRSLHEAGIEVILDVV
MMAR_2379|M.marinum_M RNYWGYNTFGFFAPHNQYAANRQAGSAVGEFKSMVRSLHEAGIEVILDVV
MUL_1555|M.ulcerans_Agy99 RNYWGYNTFGFFAPHNQYAANRQAGSAVGEFKSMVRSLHEAGIEVILDVV
MAV_3210|M.avium_104 RNYWGYNTFGFFAPHNQYAANRNS--SVAEFKSMVRSFHEAGIEVILDVV
MAB_2690|M.abscessus_ATCC_1997 RNYWGYNTFGYLAPHAGYASSPHAGGAVAEFKAMVRALHEAGIEVILDVV
TH_1798|M.thermoresistible__bu RNYWGYNTFGFFAPHNEYAATGASG-AVAEFKTMVKTFHEAGIEVILDVV
********.*::*** **:. : :*.***:**: :*:**********
Mflv_3631|M.gilvum_PYR-GCK YNHTAEGNHLGPTVNFRGIDNAAYYRLLDGDLRYYKDFTGTGNSLNARHP
Mvan_2786|M.vanbaalenii_PYR-1 YNHTAEGNHLGPTINFRGIDNAAYYRLLDGDLRLYKDFTGTGNSLNARHP
MSMEG_3186|M.smegmatis_MC2_155 YNHTAEGNHLGPTINFRGIDNAAYYRLLDGQPEFYKDFTGTGNSLNARHP
Mb1591c|M.bovis_AF2122/97 YNHTAEGNHLGPTINFRGIDNTAYYRLMDHDLRFYKDFTGTGNSLNARHP
Rv1564c|M.tuberculosis_H37Rv YNHTAEGNHLGPTINFRGIDNTAYYRLMDHDLRFYKDFTGTGNSLNARHP
MMAR_2379|M.marinum_M YNHTAEGNRLGPTINFRGIDNSAYYRLVDSDLRWYKDYTGTGNSLNARHP
MUL_1555|M.ulcerans_Agy99 YNHTAEGNRLGPTINFRGIDNSAYYRLVDSDLRWYKDYTGTGNSLNARHP
MAV_3210|M.avium_104 YNHTAEGNHLGPTINFRGIDNAAYYRLVDTDLRRYKDYTGTGNSLNPRHP
MAB_2690|M.abscessus_ATCC_1997 YNHTAEGDHIGPTLSFRGIDNRAYYKLNDDNLARYTDYTGTGNSLNARNP
TH_1798|M.thermoresistible__bu YNHTAEGNHLGPTINFRGIDNAAYYRLLDEDPRYYKDFTGTGNSLNARHP
*******:::***:.****** ***:* * : *.*:********.*:*
Mflv_3631|M.gilvum_PYR-GCK HTLQLIMDSLRYWVLDMHVDGFRFDLASTLAREFYDVDRLSAFFDIIQQD
Mvan_2786|M.vanbaalenii_PYR-1 HTLQLIMDSLRYWVLDMHVDGFRFDLASTLAREFYDVDRLSAFFDIIQQD
MSMEG_3186|M.smegmatis_MC2_155 HTLQLIMDSLRYWVLDMHVDGFRFDLASTLAREFYDVDRLSAFFDLVQQD
Mb1591c|M.bovis_AF2122/97 HTLQLIMDSLRYWVIEMHVDGFRFDLASTLARELHDVDRLSAFFDLVQQD
Rv1564c|M.tuberculosis_H37Rv HTLQLIMDSLRYWVIEMHVDGFRFDLASTLARELHDVDRLSAFFDLVQQD
MMAR_2379|M.marinum_M HVLQLIMDSLRYWVIEMHVDGFRFDLAATLARELHDVDRLSAFFDLVQQD
MUL_1555|M.ulcerans_Agy99 HVLQLIMDSLRYWVIEMHVDGFRFDLAATLARELHDVDRLSAFFDLVQQD
MAV_3210|M.avium_104 HVLQLIMDSLRYWVTEMHVDGFRFDLAATLARELHDVDRLSAFFDLVQQD
MAB_2690|M.abscessus_ATCC_1997 HTLQLIMDSLRYWVTEMHVDGFRFDLASTLARELHDVDRLSAFFDLVQQD
TH_1798|M.thermoresistible__bu HTLQLIMDSLRYWVTDMHVDGFRFDLAATLARELYDVDRLSGFFDLVQQD
*.************ :***********:*****::******.***::***
Mflv_3631|M.gilvum_PYR-GCK PVISQVKLIAEPWDIGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
Mvan_2786|M.vanbaalenii_PYR-1 PVISQVKLIAEPWDIGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
MSMEG_3186|M.smegmatis_MC2_155 PVVSQVKLIAEPWDVGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
Mb1591c|M.bovis_AF2122/97 PVVSQVKLIAEPWDVGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
Rv1564c|M.tuberculosis_H37Rv PVVSQVKLIAEPWDVGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
MMAR_2379|M.marinum_M PVVSQVKLIAEPWDVGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
MUL_1555|M.ulcerans_Agy99 PVVSQAKLIAEPWDVGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
MAV_3210|M.avium_104 PIVSQVKLIAEPWDVGEGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
MAB_2690|M.abscessus_ATCC_1997 PIVSQVKLIAEPWDIGEGGYQVGNFPGLWTEWNGKFRDTVRDYWRGQPAT
TH_1798|M.thermoresistible__bu PVVSQVKLIAEPWDVGPGGYQVGNFPGLWTEWNGKYRDTVRDYWRGEPAT
*::**.********:* ******************:**********:***
Mflv_3631|M.gilvum_PYR-GCK LGEFASRLTGSSDLYEATGRRPSASINFVTCHDGFTLNDLVSYNEKHNEA
Mvan_2786|M.vanbaalenii_PYR-1 LGEFASRLTGSSDLYEATGRRPSASINFVTCHDGFTLNDLVSYNEKHNEA
MSMEG_3186|M.smegmatis_MC2_155 LGEFASRLTGSSDLYEATGRRPSASINFVTCHDGFTLNDLVSYNEKHNEA
Mb1591c|M.bovis_AF2122/97 LGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLNDLVSYNDKHNEA
Rv1564c|M.tuberculosis_H37Rv LGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLNDLVSYNDKHNEA
MMAR_2379|M.marinum_M LGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLNDLVSYNEKHNSA
MUL_1555|M.ulcerans_Agy99 LGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLNDLVSYNEKHNSA
MAV_3210|M.avium_104 LGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLNDLVSYNEKHNMA
MAB_2690|M.abscessus_ATCC_1997 LGEFASRLTGSSDLYEATGRRPSASINFVTAHDGFTLRDLVSYNEKHNEA
TH_1798|M.thermoresistible__bu LGEFASRLTGSSDLYEATGRRPWASINFVTCHDGFTLADLVSYNEKHNEA
********************** *******.****** ******:*** *
Mflv_3631|M.gilvum_PYR-GCK NGEDNRDGESHNRSWNCGVEGPTDDPDITALRAKQMRNILATLMLSQGTP
Mvan_2786|M.vanbaalenii_PYR-1 NGEDNRDGESHNRSWNCGVEGPTDDEEILALRGKQMRNIMATLMLSQGTP
MSMEG_3186|M.smegmatis_MC2_155 NGEDNRDGESHNRSWNCGVEGPTDDPEILALRAKQMRNIMATLMLSQGTP
Mb1591c|M.bovis_AF2122/97 NGENNRDGESYNRSWNCGVEGPTDDPDILALRARQMRNMWATLMVSQGTP
Rv1564c|M.tuberculosis_H37Rv NGENNRDGESYNRSWNCGVEGPTDDPDILALRARQMRNMWATLMVSQGTP
MMAR_2379|M.marinum_M NGEDNRDGESHNRSWNCGIEGPTDDPDIVALRYRQMRNFWATLMVSQGTP
MUL_1555|M.ulcerans_Agy99 NGEDNRDGERHNRSWNCGIEGPTDDPDIVALRYRQMRNFWATLMVSQGTP
MAV_3210|M.avium_104 NGEDNRDGESHNRSWNCGVEGPTDDPDITELRYRQMRNFWATLMVSQGTP
MAB_2690|M.abscessus_ATCC_1997 NGENNQDGETYNRSWNCGVEGPTDDPQILALRARQMRNIFATLVLSQGTP
TH_1798|M.thermoresistible__bu NGEDNNDGESHNRSWNCGAEGPTDDPQVNALRARQQRNFLATLLLSQGVP
***:*.*** :******* ****** :: ** :* **: ***::***.*
Mflv_3631|M.gilvum_PYR-GCK MIAHGDEMGRTQQGNNNVYCQDSELSWMDWSLCEKNADLVEFTRKVVSFR
Mvan_2786|M.vanbaalenii_PYR-1 MIAHGDEMGRTQRGNNNVYCQDSELSWMDWSLCETNADLVEFTRRMVKFR
MSMEG_3186|M.smegmatis_MC2_155 MIAHGDEIGRTQLGNNNVYCQDSELSWMDWSLCETNADHLEFTRKVVAFR
Mb1591c|M.bovis_AF2122/97 MIAHGDEIGRTQYGNNNVYCQDSELSWMDWSLVDKNADLLAFARKATTLR
Rv1564c|M.tuberculosis_H37Rv MIAHGDEIGRTQYGNNNVYCQDSELSWMDWSLVDKNADLLAFARKATTLR
MMAR_2379|M.marinum_M MIAHGDEIGRTQQGNNNVYCQDSEISWMDWSLVEANADLLAFARKVTTLR
MUL_1555|M.ulcerans_Agy99 MIAHGDEIGRTQQGNNNVYCQDSEISWMDWSLVEANADLLAFARKVTTLR
MAV_3210|M.avium_104 MIAHGDEFGRTQNGNNNVYCQDSELSWMDWSLVDKNSDLLAFARRATTLR
MAB_2690|M.abscessus_ATCC_1997 MLSHGDEIGRTQQGNNNVYCQDSALSWMDWELATANADLLQFARSVIALR
TH_1798|M.thermoresistible__bu MLCHGDELGRTQQGNNNVYCQDNELSWIDWSTADT--ELIEFTRTVSALR
*:.****:**** *********. :**:**. : : *:* :*
Mflv_3631|M.gilvum_PYR-GCK KKHPVFRRRRFFEGKPLR--TGEQIRDIAWFNPSGREMTSEDWGSGF-DC
Mvan_2786|M.vanbaalenii_PYR-1 KDHPAFRRRRFFEGKPQS--TGEQIRDITWMNPSGTEMTQEDWGSGF-EC
MSMEG_3186|M.smegmatis_MC2_155 KQHPVFRRRRFFEGKPIR--SGDQVRDIAWLTRAGTEMTPEDWGAGLGTC
Mb1591c|M.bovis_AF2122/97 KNHKVFRRRRFFEGEPIR--SGDEVRDIAWLTPSGREMTHEDWGRGFDRC
Rv1564c|M.tuberculosis_H37Rv KNHKVFRRRRFFEGEPIR--SGDEVRDIAWLTPSGREMTHEDWGRGFDRC
MMAR_2379|M.marinum_M RNHPVFRRRRFFEGEPIR--TGDEVRDIAWLTPSGQEMLHDDWNRSFHKC
MUL_1555|M.ulcerans_Agy99 RNHPVFRRRRFFEGEPIR--TGDEVRDIAWLTPSGQEMLHDDWNRSFHKC
MAV_3210|M.avium_104 TKHPVFRRRRFFEGEPIR--SGDEVRDIAWLTPGGREMTHEDWGQSFHKC
MAB_2690|M.abscessus_ATCC_1997 KQHPVFRRRRFFAGRPIR--EGEEVRDIAWLTPAGEEMTTADWDSGFGKS
TH_1798|M.thermoresistible__bu TAHPVFRRRRFFSGRPVRGHGGDRLPDIAWLAPDASEMTDEDWESGYAKS
* .******* *.* *:.: **:*: . ** ** . .
Mflv_3631|M.gilvum_PYR-GCK VSVFLNGDAIPAPDERGQRVSDDSFLLCFNAHDEPLEFVAPDGDYANEWA
Mvan_2786|M.vanbaalenii_PYR-1 VSVFLNGDAIPSPDERGERITDDSFLLCFNAHDEPLDFVLPNADYAKEWT
MSMEG_3186|M.smegmatis_MC2_155 VAVFLNGEAIPTPDARGERVVDDSFLLCFNSNDHPEDFVTPNGDYAAEWT
Mb1591c|M.bovis_AF2122/97 VAVFLNGEAITAPDARGERVVDDSFLLCFNAHDHDVEFVMPHDGYAQQWT
Rv1564c|M.tuberculosis_H37Rv VAVFLNGEAITAPDARGERVVDDSFLLCFNAHDHDVEFVMPHDGYAQQWT
MMAR_2379|M.marinum_M VAVFLNGEAITAPNARGERVVDDSFLLCFNAHDETVEFVMPHDDYANEWT
MUL_1555|M.ulcerans_Agy99 VAVFLNGEAITAPNARGERVVDDSFLLCFNAHDETVEFVMPHDDYANEWT
MAV_3210|M.avium_104 VAVFLNGDAITAPNARGERVVDDSFLLCFNAGEQPVQFVMPGGDYAKEWT
MAB_2690|M.abscessus_ATCC_1997 LAVFLNGDAIPEPNARGERVSGDSFLLCFNAYDEPLDFVTPDGDYATQWT
TH_1798|M.thermoresistible__bu MAVFLNGQGIPDLDVRGQRVVDDSFLLCFNAHFEPIEFTLPPPDFAGSWL
::*****:.*. : **:*: .********: . :*. * .:* .*
Mflv_3631|M.gilvum_PYR-GCK GAIDTADRQG----ESTLTVVAGQTISVDPRSLLVLRKTA---
Mvan_2786|M.vanbaalenii_PYR-1 GAVDTTHPEG----ASTLEVAAGETISLQPRSLLVLRKTA---
MSMEG_3186|M.smegmatis_MC2_155 ADLDTADPVG----NSDLVVKAGEKISIQPRSVLVLRKTA---
Mb1591c|M.bovis_AF2122/97 GELDTNDPVG----DIDLTVTATDTFSVPARSLLVLRKTL---
Rv1564c|M.tuberculosis_H37Rv GELDTNDPVG----DIDLTVTATDTFSVPARSLLVLRKTL---
MMAR_2379|M.marinum_M VELDTNHPAG----DADAVVKAEDTVSVPGRSLILLRKTA---
MUL_1555|M.ulcerans_Agy99 VELDTNHPAG----DADAVVKAEDTVSVPGRSLILLRKTA---
MAV_3210|M.avium_104 VELDTNEPTGRKEGAEPLVVHAEEELTLPSRSLLILRKTL---
MAB_2690|M.abscessus_ATCC_1997 AVLDTATPNG----QCSTVVKAGEAVRVQDRALVVLRKSG---
TH_1798|M.thermoresistible__bu RVIDTAEPAG----PAAEPLDAGAPLSVEGRAVVVLQAVPAGD
:** * : * . : *::::*: