For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFTRRFAASMVGTTLTAATLGLAALGFAGTASASSTDEAFLAQLQADGITPPSAARAIKDAHAVCDALDE GHSAKAVIKAVAKATGLSAKGAKTFAVDAASAYCPQYVTSS
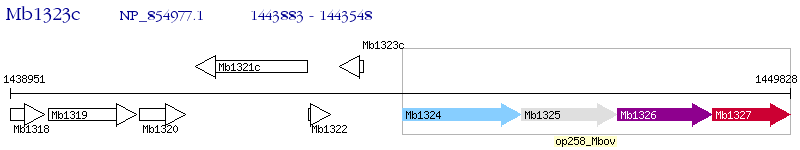
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1323c | - | - | 100% (111) | hypothetical protein Mb1323c |
| M. bovis AF2122 / 97 | Mb1302c | - | 8e-16 | 42.27% (97) | hypothetical protein Mb1302c |
| M. bovis AF2122 / 97 | Mb3389 | - | 7e-09 | 37.00% (100) | hypothetical protein Mb3389 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5185 | - | 9e-09 | 37.74% (106) | hypothetical protein Mflv_5185 |
| M. tuberculosis H37Rv | Rv1291c | - | 2e-57 | 100.00% (111) | hypothetical protein Rv1291c |
| M. leprae Br4923 | MLBr_00676 | - | 3e-06 | 30.43% (92) | hypothetical protein MLBr_00676 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4121 | - | 1e-46 | 81.42% (113) | hypothetical protein MMAR_4121 |
| M. avium 104 | MAV_1419 | - | 1e-15 | 40.00% (110) | hypothetical protein MAV_1419 |
| M. smegmatis MC2 155 | MSMEG_3681 | - | 2e-05 | 33.75% (80) | hypothetical protein MSMEG_3681 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3987 | - | 2e-46 | 80.53% (113) | hypothetical protein MUL_3987 |
| M. vanbaalenii PYR-1 | Mvan_4716 | - | 1e-07 | 31.63% (98) | hypothetical protein Mvan_4716 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1323c|M.bovis_AF2122/97 MFTRRFAASMVGTTLTAATLGLAA--------------------------
Rv1291c|M.tuberculosis_H37Rv MFTRRFAASMVGTTLTAATLGLAA--------------------------
MMAR_4121|M.marinum_M MFTRRFAASLAGTTLTAASLGLAA--------------------------
MUL_3987|M.ulcerans_Agy99 MFTRRFAASLAGTTLTAASLGLAA--------------------------
MAV_1419|M.avium_104 MFSRRIIT-----VLITAAIGLAA--------------------------
Mflv_5185|M.gilvum_PYR-GCK MFAR-----------RAASAGIAA--------------------------
Mvan_4716|M.vanbaalenii_PYR-1 MRGK---------LIVSAAVAAAG--------------------------
MLBr_00676|M.leprae_Br4923 MFLRLMLSALKALQRLGAVMNSLARIDHWIWLFRCQPLTIRLLVATAALF
MSMEG_3681|M.smegmatis_MC2_155 MMAGLMGSMIGAAPALADPADPAVPG----------------------IP
*
Mb1323c|M.bovis_AF2122/97 LGFAG--TASASST-DEAFLAQLQADGITPPSAARAIKDAHAVCDALDEG
Rv1291c|M.tuberculosis_H37Rv LGFAG--TASASST-DEAFLAQLQADGITPPSAARAIKDAHAVCDALDEG
MMAR_4121|M.marinum_M LGFAATATASAGST-DDAFLAQIQADGITPPSAARAISDAHAVCTALDEG
MUL_3987|M.ulcerans_Agy99 LGFAATATATAGST-DDAFLAQIQADGITPPSAARAISDAHAVCTALDEG
MAV_1419|M.avium_104 VGLAGPAAARTVQTGDEMFLAQMRSLGVSFPSPQEAVREGHQVCAELSAG
Mflv_5185|M.gilvum_PYR-GCK VAAVGLAAPASASPEVAVFLDRLQKVGITSSNPYATIYDAYAVCRELDRG
Mvan_4716|M.vanbaalenii_PYR-1 LATALAAPAFADET-DDIFISALQNQGIPFSTAENAIELASAVCDYAAAG
MLBr_00676|M.leprae_Br4923 TAATAFEVPAEADAIDDTFIKALNHAGVNFGEPRSAMTMGHYVCPILAKS
MSMEG_3681|M.smegmatis_MC2_155 MDPAVPVPGAPGAPTDQAFVDALGQAGVNAVDPAQAAAMGEAVCPMLAEQ
. . *: : *: . : . **
Mb1323c|M.bovis_AF2122/97 HSAKAVIKAVAKATG-LSAKGAKTFAVDAASAYCPQYVTSS---------
Rv1291c|M.tuberculosis_H37Rv HSAKAVIKAVAKATG-LSAKGAKTFAVDAASAYCPQYVTSS---------
MMAR_4121|M.marinum_M NSPTTVINAVAESTG-LSSKGAKTFAIDAASAYCPEYVTSS---------
MUL_3987|M.ulcerans_Agy99 NSPTTVINAVAESTG-LSSKGAKTFAIDAASAYCPEYVTSS---------
MAV_1419|M.avium_104 KTPTAVTVAVFRQTN-LTPPQAAGLVTAATQAYCPQFSGQPA--------
Mflv_5185|M.gilvum_PYR-GCK TSPTQVVGFVLGDNPDLDWEAAADYVVLANMTYCPPV-------------
Mvan_4716|M.vanbaalenii_PYR-1 QDPTQIALEIMEPAG-WTAEQSGFFVGAATQSYCPS--------------
MLBr_00676|M.leprae_Br4923 GGNFAAAVQRIRGNSDMSPQMAETFAKIAISIYCPTMMANVASGNLPSLP
MSMEG_3681|M.smegmatis_MC2_155 GQSAADVASTVADTGGMPLGPATMFTGIAVSMFCPAMVSRLSQGVPPDLP
: . * :**
Mb1323c|M.bovis_AF2122/97 ----------
Rv1291c|M.tuberculosis_H37Rv ----------
MMAR_4121|M.marinum_M ----------
MUL_3987|M.ulcerans_Agy99 ----------
MAV_1419|M.avium_104 ----------
Mflv_5185|M.gilvum_PYR-GCK ----------
Mvan_4716|M.vanbaalenii_PYR-1 ----------
MLBr_00676|M.leprae_Br4923 PGPGIPGI--
MSMEG_3681|M.smegmatis_MC2_155 MNLPWPMFGF