For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGESAESGSRQLPAMSPPRRSVAYRRKIVDALWWAACVCCLAVVITPTLWMLIGVVSRAVPVFHWSVLVQ DSQGNGGGLRNAIIGTAVLAIGVILVGGTVSVLTGIYLSEFATGKTRSILRGAYEVLSGIPSIVLGYVGY LALVVYFDWGFSLAAGVLVLSVMSIPYIAKATESALAQVPTSYREAAEALGLPAGWALRKIVLKTAMPGI VTGMLVALALAIGETAPLLYTAGWSNSPPTGQLTDSPVGYLTYPIWTFYNQPSKSAQDLSYDAALLLIVF LLLLIFIGRLINWLSRRRWDV
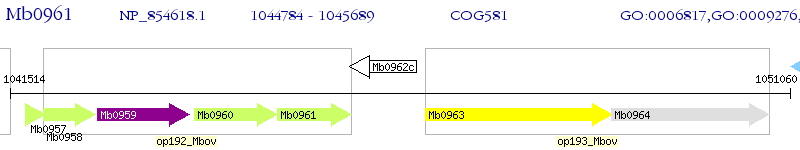
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0961 | pstA2 | - | 100% (301) | phosphate ABC transporter transmembrane protein |
| M. bovis AF2122 / 97 | Mb0953 | pstA1 | 7e-28 | 28.71% (303) | phosphate ABC transporter transmembrane protein |
| M. bovis AF2122 / 97 | Mb0952 | pstC2 | 9e-12 | 24.68% (231) | phosphate ABC transporter transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1663 | - | 6e-27 | 28.37% (289) | phosphate ABC transporter, inner membrane subunit PstA |
| M. tuberculosis H37Rv | Rv0936 | pstA2 | 1e-171 | 100.00% (301) | phosphate ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 2e-28 | 28.77% (292) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_4850c | - | 3e-26 | 28.47% (295) | phosphate ABC transporter permease |
| M. marinum M | MMAR_4578 | pstA1 | 2e-28 | 29.90% (301) | phosphate-transport integral membrane ABC transporter PstA1 |
| M. avium 104 | MAV_0766 | pstA | 3e-26 | 30.14% (292) | phosphate ABC transporter, permease protein PstA |
| M. smegmatis MC2 155 | MSMEG_5780 | pstA | 1e-29 | 31.23% (285) | phosphate ABC transporter, permease protein PstA |
| M. thermoresistible (build 8) | TH_0381 | pstA | 4e-30 | 32.25% (276) | phosphate ABC transporter, permease protein PstA |
| M. ulcerans Agy99 | MUL_4439 | pstA1 | 1e-28 | 29.90% (301) | phosphate-transport integral membrane ABC transporter PstA1 |
| M. vanbaalenii PYR-1 | Mvan_5090 | - | 9e-27 | 28.37% (289) | phosphate ABC transporter, inner membrane subunit PstA |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4578|M.marinum_M ----MTTNALDQPVKVVVFRPLSLVRRIKNSVATVLFFASFAVALVPLVW
MUL_4439|M.ulcerans_Agy99 ----MTTNALDEPVKVVVFRPLSLVRRIKNSVAAVLLFASFAVALVPLVW
MLBr_02093|M.leprae_Br4923 ----MNLDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSFLIALVPLVW
MAB_4850c|M.abscessus_ATCC_199 -----MTSTLDEPIKTSTFHSVSTVRRIKNRVATTLFATAFGVALVPLAW
Mflv_1663|M.gilvum_PYR-GCK -----MTTTLEKAVKGPTFHPVSASRKFKNGLATTLFAGSFVVAMIPLVW
Mvan_5090|M.vanbaalenii_PYR-1 -----MTATLDEAVKGPTFHPLSAARKFKNGLATTLFTASFVVAMIPLVW
MSMEG_5780|M.smegmatis_MC2_155 -MTTSMTSTLEQPVKAPAFQGVSLRRRLTDHTATVLVTTSVLIALIPLLW
TH_0381|M.thermoresistible__bu MATPTTTAELDAPVKPTTFQPVSLRRKAVNHMATVLVTASVLIALIPLVW
MAV_0766|M.avium_104 -----MTSMLDRPLKSRTFSPLSRRRRAANSVATVLVSVSLLVAVTPLVM
Mb0961|M.bovis_AF2122/97 MGESAESGSRQLPAMSPPRRSVAYRRKIVDALWWAACVCCLAVVITPTLW
Rv0936|M.tuberculosis_H37Rv MGESAESGSRQLPAMSPPRRSVAYRRKIVDALWWAACVCCLAVVITPTLW
: . :: *: : .. :.: *
MMAR_4578|M.marinum_M LLWVVIERGWYAVVQPDWWTHSLRGVLPEEFAGGVYHALYGTLVQAGIAA
MUL_4439|M.ulcerans_Agy99 LLWVVIERGWYAVVQPDWWTHSLRGVLPEEFAGGVYHALYGTLVQAGIAA
MLBr_02093|M.leprae_Br4923 LLSVVVARGFYAVTRFDWWTHSLRGVMPEEFAGGVYHALYGSLVQAVVAA
MAB_4850c|M.abscessus_ATCC_199 VLFIVLERGWQAVTTSGWWTRSLQGVLPEEFAGGIYHALYGTLVQAGIAA
Mflv_1663|M.gilvum_PYR-GCK LLYTVVAKGISAIGSSTWWMNSLAGVLPEEFAGGVYHAIYGTLIQAAIAA
Mvan_5090|M.vanbaalenii_PYR-1 LIYTVLAKGVAAIVSSTWWTNSLAGVLPEEFAGGVYHAIYGTLIQAAIAA
MSMEG_5780|M.smegmatis_MC2_155 VLYSVITKGIHAVTSPTWFTNSQAGMTAFSPGGGVYHAIVGTVLQGLVCS
TH_0381|M.thermoresistible__bu VLVSVFVKGIGVVTNTTWWTHSQAGMTAFAAGGGAYHAIVGTLLQGLVCA
MAV_0766|M.avium_104 VLCSVVVKGFRAITSTVWWSHSQAGMTAFVTGGGAYHALVGTVLQGLVCA
Mb0961|M.bovis_AF2122/97 MLIGVVSRAVPVFHWSVLVQDSQG------NGGGLRNAIIGTAVLAIGVI
Rv0936|M.tuberculosis_H37Rv MLIGVVSRAVPVFHWSVLVQDSQG------NGGGLRNAIIGTAVLAIGVI
:: *. :. .. * .** :*: *: : .
MMAR_4578|M.marinum_M LLAVPLGLMTAIYLVEYGSG-RLARLTTFMVDVLAGVPSIVAALFIFSLW
MUL_4439|M.ulcerans_Agy99 LLAVPLGLMTAIYLVEYGSG-RLARLTTFMVDVLAGVPSIVAALFIFSLW
MLBr_02093|M.leprae_Br4923 IMAVPLGSMTAVYLVEYGSG-PFVRVTTFMVDVLAGVPSIVAALFIFCLW
MAB_4850c|M.abscessus_ATCC_199 VLAIPLGVMAAIYLVEYGRG-TLAKLTTFMVDVLAGVPSIVAALFIFALW
Mflv_1663|M.gilvum_PYR-GCK AISVPLGVMAAVFLVEYGGG-RLAKVTTFMVDILAGVPSIVAALFVFALW
Mvan_5090|M.vanbaalenii_PYR-1 LLSVPLGVMAAVYLVEYGQG-RFAKVTTFMVDILAGVPSIVAALFIFALW
MSMEG_5780|M.smegmatis_MC2_155 LISIPIGVMVAVYLVEYAGGSRLGKITTFMVDILTGVPSIVAALFIYALW
TH_0381|M.thermoresistible__bu AISIPIGVFVGIYLVEYGAGTRLGKLTTFMVDILTGVPSIVAALFIYALW
MAV_0766|M.avium_104 AISIPIGLMVAIYLVEYGGGTPLGRLASFMVDILSGVPSIVAALFVYALC
Mb0961|M.bovis_AF2122/97 LVGGTVSVLTGIYLSEFATG-KTRSILRGAYEVLSGIPSIVLGYVGYLAL
Rv0936|M.tuberculosis_H37Rv LVGGTVSVLTGIYLSEFATG-KTRSILRGAYEVLSGIPSIVLGYVGYLAL
:. .:. :..::* *:. * : ::*:*:**** . . :
MMAR_4578|M.marinum_M IATLGFQQSALAVSLALVLLMLPVVVRSTEEMLRLVPDELREASFALGVP
MUL_4439|M.ulcerans_Agy99 IATLGFQQSALAVSLALVLLMLPVVVRSTEEMLRLVPDELREASFALGVP
MLBr_02093|M.leprae_Br4923 IATLGFQQSAFAVSLALVLLMLPVVVRSTEEILRLVPDELREACYALGIP
MAB_4850c|M.abscessus_ATCC_199 IATLGFPQSALAVSLALVLLMLPVVVRSTEEMLKLVPDELREASYALGIA
Mflv_1663|M.gilvum_PYR-GCK IATLGFEQSAFAVSLALVLLMLPVVVRNTEEMLKLVPDELREASYALGIP
Mvan_5090|M.vanbaalenii_PYR-1 IATLGFEQSAFAVSLALVLLMLPVVVRNTEEMLKLVPDELREASYALGVP
MSMEG_5780|M.smegmatis_MC2_155 VATLGFQRSGFAVSLSLVLLMIPVIVRATEEMLRIVPMDLREASYALGVP
TH_0381|M.thermoresistible__bu VATLGFGRSGLAVSLSLVLLMVPVIVRSTEEMLRIVPMDLREAAYALGVP
MAV_0766|M.avium_104 VATLGLPRSEFAVSLALVLLMLPVIVRATEEMLRIVPVDLREASYALGIT
Mb0961|M.bovis_AF2122/97 VVYFDWGFSLAAGVLVLSVMSIPYIAKATESALAQVPTSYREAAEALGLP
Rv0936|M.tuberculosis_H37Rv VVYFDWGFSLAAGVLVLSVMSIPYIAKATESALAQVPTSYREAAEALGLP
:. :. * * * * :: :* :.: **. * ** . ***. ***:.
MMAR_4578|M.marinum_M KWKTIVRIVVPIAMPGIVSGVLLSVARVIGETAPVLVLVGYSRSINFDVL
MUL_4439|M.ulcerans_Agy99 KWKTIVRIVVPIAMPGIVSGVLLSVARVIGETAPVLVLVGYSRSINFDVL
MLBr_02093|M.leprae_Br4923 KWKTIVRIVFPIATPGIISGVLLSIARVIGETAPVLVLVGYSRSINFDIF
MAB_4850c|M.abscessus_ATCC_199 KWKTIVKVVVPTALPGIISGILLSVARVIGETAPVLVLVGYARSINFDVF
Mflv_1663|M.gilvum_PYR-GCK KWKTIVRIVVPTALPGMISGILLALARVMGETAPVLVLVGYSRSINMDAF
Mvan_5090|M.vanbaalenii_PYR-1 KWKTIARIVVPTALPGMISGILLALARVMGETAPVLVLVGYSRSINMDAF
MSMEG_5780|M.smegmatis_MC2_155 KWKTIVRIVIPTALSGIVTGVMLSLARVMGETAPLLVLVGYAQAMNFDIF
TH_0381|M.thermoresistible__bu KWKTIVRIVIPTALSGIVTGIMLALARVMGETAPLLVLVGYAQAMNFDMF
MAV_0766|M.avium_104 KWKTIARVVLPTGLSGIVTGILLAMARVMGETAPLLILVGYAQSMNFDIF
Mb0961|M.bovis_AF2122/97 AGWALRKIVLKTAMPGIVTGMLVALALAIGETAPLLYTAGWSNSPPTGQL
Rv0936|M.tuberculosis_H37Rv AGWALRKIVLKTAMPGIVTGMLVALALAIGETAPLLYTAGWSNSPPTGQL
:: ::*. . .*:::*:::::* .:*****:* .*::.: . :
MMAR_4578|M.marinum_M HG-NMASLPLLIYTELTNPEHAG---FLRVWGAALTLIIIVAAINLIAGG
MUL_4439|M.ulcerans_Agy99 HG-NMASLPLLIYTELTNPEHAG---FLRVWGAALTLIIIVAAINLIAGG
MLBr_02093|M.leprae_Br4923 DG-NMASLPLLIYTELTNPEYAG---FLRVWGAALTLIILVAGINLFGAA
MAB_4850c|M.abscessus_ATCC_199 EG-NMASLPLLIYTELINPQPAG---QMRVWGAALTLIIVVAILMALAAV
Mflv_1663|M.gilvum_PYR-GCK EG-NMASLPLLIYTELVNPEAAG---VLRVWGAALTLILIIAALYIGAAF
Mvan_5090|M.vanbaalenii_PYR-1 EG-NMASLPLLIYTELVNPEAAG---VLRVWGAALTLILIIAALYIGAAF
MSMEG_5780|M.smegmatis_MC2_155 SG-FMGSLPGMMYDQISAGAGANPVPTDRLWGAALTLILLIALLNVGARF
TH_0381|M.thermoresistible__bu TG-FMGSLPGMMYDQTSAGAGANPIPTDRLWGAALTLIVLIALLNIGARF
MAV_0766|M.avium_104 SG-FMGTLPGMMYNQASAGAGINPIPTDRLWGAALTLILVIATINIVARV
Mb0961|M.bovis_AF2122/97 TDSPVGYLTYPIWTFYNQPSKSA---QDLSYDAALLLIVFLLLLIFIGRL
Rv0936|M.tuberculosis_H37Rv TDSPVGYLTYPIWTFYNQPSKSA---QDLSYDAALLLIVFLLLLIFIGRL
. :. *. :: :.*** **:.: : .
MMAR_4578|M.marinum_M IRFLATRRR----
MUL_4439|M.ulcerans_Agy99 IRFLATRRR----
MLBr_02093|M.leprae_Br4923 FRFLATRRCSGCR
MAB_4850c|M.abscessus_ATCC_199 ATRLLTRKR----
Mflv_1663|M.gilvum_PYR-GCK INRYLTRNRI---
Mvan_5090|M.vanbaalenii_PYR-1 INRYLMRNRV---
MSMEG_5780|M.smegmatis_MC2_155 VAKIFAPKKV---
TH_0381|M.thermoresistible__bu LAKLFAPKKV---
MAV_0766|M.avium_104 ITKFLGARKS---
Mb0961|M.bovis_AF2122/97 INWLSRRRWDV--
Rv0936|M.tuberculosis_H37Rv INWLSRRRWDV--
.