For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MMARMPELSRRAVLGLGAGTVLGATSAYAIDMLLQPRTSHAAPAAAIGTNVPLAPTPALDPAPPAQAAPT MSTGSFVSAARAGKMTNWAIARPPGQTQALRPVIALHGLGGSASAVMDGGVEQGLAQAVNAGLPPFAVVS VAGGSSYWHQRASGEDAGAMVLNELIPLLDTQRLDTSRVAFLGWSMGGYGALLLGSRLGPARTAAICAVS PALWLSAGAVAPGSFDGPDDWSANSVFGLPALGSIPIRVDCGNSDPFYAATKQFVAQLPHPPAGGFSPGG HNGGFWSAQLPAELTWFAPLLTG
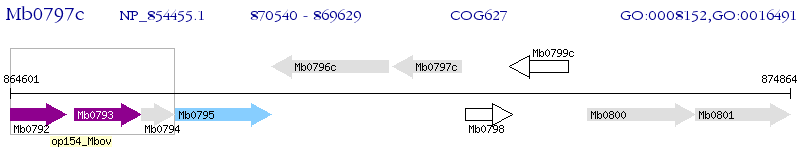
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0797c | - | - | 100% (303) | hypothetical protein Mb0797c |
| M. bovis AF2122 / 97 | Mb0532c | - | 2e-93 | 56.71% (298) | hypothetical protein Mb0532c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1609 | - | 1e-100 | 62.29% (297) | putative esterase |
| M. tuberculosis H37Rv | Rv0774c | - | 1e-175 | 99.34% (303) | hypothetical protein Rv0774c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0684c | - | 1e-100 | 62.42% (298) | hypothetical protein MAB_0684c |
| M. marinum M | MMAR_4919 | - | 1e-146 | 83.06% (301) | hypothetical protein MMAR_4919 |
| M. avium 104 | MAV_0719 | - | 1e-140 | 79.34% (305) | putative esterase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_5851 | - | 1e-118 | 70.67% (300) | putative esterase superfamily protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0487 | - | 1e-145 | 82.06% (301) | hypothetical protein MUL_0487 |
| M. vanbaalenii PYR-1 | Mvan_5149 | - | 1e-100 | 62.46% (301) | putative esterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0797c|M.bovis_AF2122/97 ---------MMARMPELSRRAVLGLGAGTVLGATSAYAIDMLLQPRTSHA
Rv0774c|M.tuberculosis_H37Rv ---------MMARMPELSRRAVLGLGAGTVLGATSAYAIDMLLQPRTSHA
MMAR_4919|M.marinum_M ---------MMARMPDLSRRAVLGLGASATLGAVGAYALDMALLPRTSRA
MUL_0487|M.ulcerans_Agy99 MPFSSNVHGMMARMPDLSRRAVLGLGASATLGAVGAYALDMALLPRTSRA
MAV_0719|M.avium_104 ---------MMTRMPDLSRRAVLRLGAGAAVGAVGAYGLDILLQPRASQA
MSMEG_5851|M.smegmatis_MC2_155 ----------MAGMPTLSRRAVLRLGVGAAAGAAGAVAFGVAATATPGQP
MAB_0684c|M.abscessus_ATCC_199 -------------MRDLSRRSLLKLGAATGAGAA-------LLGAG----
Mflv_1609|M.gilvum_PYR-GCK -------------MPMINRRDVLSLGLRAAMVATAGTAAASLLDTPAS--
Mvan_5149|M.vanbaalenii_PYR-1 ---------MMSRMPTLNRREMLALGLRAGLAATA----ASALGTAVA--
* :.** :* ** : *. .
Mb0797c|M.bovis_AF2122/97 APAAAIGT---NVPLAPTPALDPAPPAQAAPTMSTGSFVSAARAGKMTNW
Rv0774c|M.tuberculosis_H37Rv APAAAIGT---NVPLAPTPALDPAPPAQAAPTMSTGSFVSAARAGKMTNW
MMAR_4919|M.marinum_M APAPPIGT---NVPLAPARPLDPPPPAQAAPTMTTGSFVSAARGGITTNW
MUL_0487|M.ulcerans_Agy99 APAPPIGT---NVPLAPARPLDPPPPAQAAPTMTTGSFVSAARGGITTNW
MAV_0719|M.avium_104 MPAAGTGTPLAPPPPAPGPPRDPAPPAAAAPTMVTGSFVSAARGGVNTNW
MSMEG_5851|M.smegmatis_MC2_155 APQTPPAP------QAPPVPLEP--PATAAPTYVTGSFVSAARGGVATNW
MAB_0684c|M.abscessus_ATCC_199 -----------------------AFPAGADGAIAAGSFVSAARGGVRTEW
Mflv_1609|M.gilvum_PYR-GCK -------------------------AAPPAPTYTTGSFPSAARGGAVTNW
Mvan_5149|M.vanbaalenii_PYR-1 -------------------------SAAPAPTFVTGSFASAARGGAVTNW
.* . : :*** ****.* *:*
Mb0797c|M.bovis_AF2122/97 AIARPPGQTQALRPVIALHGLGGSASAVMDGGVEQGLAQAVNAGLPPFAV
Rv0774c|M.tuberculosis_H37Rv AIARPPGQTQALRPVIALHGLGGSASAVMDGGVEQGLAQAVNAGLPPFAV
MMAR_4919|M.marinum_M AIARPPGQTQPLRPVIALHGKGSDAATVMAGGVEQGLAQAVNAGLPPFAV
MUL_0487|M.ulcerans_Agy99 AIARPPGQTQPLRPVIALHGKGSDAATVMAGGVEQGLAQAVNAALPPFAV
MAV_0719|M.avium_104 AIARPPGQTKPLRPVIALHGKGSDAATVMAGGVEQGLAQAVNAGLPPFAV
MSMEG_5851|M.smegmatis_MC2_155 AIARPPGQTAPLRPVIALHGKGQDAAGVMAGGVEQGLAEAVAAGLPPFAV
MAB_0684c|M.abscessus_ATCC_199 RIARPPGVTGPIRPIIALHGKGQTAAGVMEGGVENMLAQAVAAGAKPFAL
Mflv_1609|M.gilvum_PYR-GCK AIARPPGRTEPLRPVILLHGKDSSAATVMSMGVEQFLADAVRAGLPPFAM
Mvan_5149|M.vanbaalenii_PYR-1 VIARPPDQTAPLRPVILLHGKDSSAEMVMSMGVERFLADAVRAGLPPFAM
*****. * .:**:* *** . * ** ***. **:** *. ***:
Mb0797c|M.bovis_AF2122/97 VSVAGGSSYWHQRASGEDAGAMVLNELIPLLDTQRLDTSRVAFLGWSMGG
Rv0774c|M.tuberculosis_H37Rv VSVDGGSSYWHQRASGEDAGAMVLNELIPLLDTQRLDTSRVAFLGWSMGG
MMAR_4919|M.marinum_M VAVDGGGSYWHKRASGEDSGAMVLDELIPMLDSQRLDTSRVAFLGWSMGG
MUL_0487|M.ulcerans_Agy99 VAVDGGGSYWHKRASGEDSGAMVLDELIPMLDSQRLDTSRVAFLGWSMGG
MAV_0719|M.avium_104 VAVDGGGSYWHKRSSGEDSGAMVLDELIPLLGNQGLDTSRVAFLGWSMGG
MSMEG_5851|M.smegmatis_MC2_155 VAVDGGGSYWHKRASGEDSGAMVLDELIPMLGEQGLDTSRVGFLGWSMGG
MAB_0684c|M.abscessus_ATCC_199 ASVDGGGGYWHKRASGEDSGAMVLNEFIPLLAEQGLDTSRVAFLGWSMGG
Mflv_1609|M.gilvum_PYR-GCK AAVDGGNGYWHRRASGDDAGAMVLDEFLPLLAAQGLDTSRVGFLGWSMGG
Mvan_5149|M.vanbaalenii_PYR-1 AAVDGGNGYWHRRASGDDPGAMVLDEFLPLLATQGLDTSRVAFLGWSMGG
.:* **..***:*:**:*.*****:*::*:* * ******.********
Mb0797c|M.bovis_AF2122/97 YGALLLGSRLGPARTAAICAVSPALWLSAGAVAPGSFDGPDDWSANSVFG
Rv0774c|M.tuberculosis_H37Rv YGALLLGSRLGPARTAAICAVSPALWLSAGSVAPGSFDGPDDWSANSVFG
MMAR_4919|M.marinum_M YGALLLGGRLGPARTAAICAVSPALWTSAGAAAPGAFDGAADYAANSVWG
MUL_0487|M.ulcerans_Agy99 YGALLLGGRLGPARTAAICAVSPALWTSAGAAAPGAFDGAADYAANSVWG
MAV_0719|M.avium_104 YGALLLGGRLGPARTAAICAVSPALWLSSGAAAPGAFDGPDDFAANSVFG
MSMEG_5851|M.smegmatis_MC2_155 YGALLLGSRLGPARTAAICAVSPALWTSPGAAAPGAFDNAEDYNANSVWG
MAB_0684c|M.abscessus_ATCC_199 YGAMLLGSRLGAGRTAAITAVSPALWTSSGAAAPGAFDGAEDYAANSVWG
Mflv_1609|M.gilvum_PYR-GCK YGALLLGSRLGAGRTAAICAVSPALWTSPGAAAPGAFDGAEDYAAHSVWG
Mvan_5149|M.vanbaalenii_PYR-1 YGAMLLGSRLGAGRTAAITAVSPALWTSPGAAAPGAFDGAADYEANSVWG
***:***.***..***** ******* *.*:.***:**.. *: *:**:*
Mb0797c|M.bovis_AF2122/97 LPALGSIPIRVDCGNSDPFYAATKQFVAQLPHPPAGGFSPGGHNGGFWSA
Rv0774c|M.tuberculosis_H37Rv LPALGSIPIRVDCGNSDPFYAATKQFVAQLPHPPAGGFSPGGHNGGFWSA
MMAR_4919|M.marinum_M MPALGSIPIRVDCGDSDPFYSATKQFIAQLPNPPAGGFSPGGHNGGFWSS
MUL_0487|M.ulcerans_Agy99 MPALGSIPIRVDCGDGDPFYSATKQFIAQLPNPPAGGFSPGGHNGGFWSS
MAV_0719|M.avium_104 MPALGSIPIRVDCGDSDPFYAATKQFIAQLPNPPAGGFSPGGHNPEFWSS
MSMEG_5851|M.smegmatis_MC2_155 QPALASIPLRIDCGTSDPFYAATRQFIAQLPNAPAGGFSPGGHNGEFWSA
MAB_0684c|M.abscessus_ATCC_199 LAALNSIPLRIDCGDSDPFYSATKQFIAQLPTPPAGGFSPGGHDGAFWSS
Mflv_1609|M.gilvum_PYR-GCK PAGLDGIPLRIDCGYDDPFASATEQFIAQLPSPPAGGFSPGGHNAQFWSG
Mvan_5149|M.vanbaalenii_PYR-1 LAALNGIPLRIDCGFDDPFYSATEQFIAQLSTPPAGGFSPGGHNAAFWSS
..* .**:*:*** .*** :**.**:***. .**********: ***.
Mb0797c|M.bovis_AF2122/97 QLPAELTWFAPLLTG
Rv0774c|M.tuberculosis_H37Rv QLPAELTWFAPLLTG
MMAR_4919|M.marinum_M QLPAELTWMAPLLVA
MUL_0487|M.ulcerans_Agy99 QLPAELTWMAPRLVA
MAV_0719|M.avium_104 QLPAELSWMAPLLTA
MSMEG_5851|M.smegmatis_MC2_155 QLPGELTWMAPLLVA
MAB_0684c|M.abscessus_ATCC_199 QIGAEMAWMAPLLTA
Mflv_1609|M.gilvum_PYR-GCK QLTSEMTWMAPLLAA
Mvan_5149|M.vanbaalenii_PYR-1 QLTSEMTWAAPLLAT
*: .*::* ** *.