For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRVDGRDIGVSGNLLQPLTRRTNDIIRAVLAAIYLVAVITSSLITRPQWVALEKSISEIVGVLSPSQSDL VYLGYGLAILALPFVILIGLIVSRQWKLLGAYAAAGLMAVLPLSISSSRIAAPRWHFDLSDRLATLLAQF LDDPRWIAILAAVLTVSGPWLPARWRHWWWALLLAFVPIHLVVSAIVPARSLLGLAVGWLVGALVVLVVG TPALEVPLDGAIRALAKRGFAVSGLAVVRPAGPGPLVLSAACEQPNAGACSEALIELYGPHQSGGGALRQ LWLKLTLRGTETAPLQASMRRAVEHRALMAIAFGDLGMANTTVIAVSPLDRGWTLYAHRPARGIGISECT KTTPTAHVWEALRTLHDQQISHGDLCSAEITVDNGAVLFGGFGEAEYGATDAQLQSDLAQLLVTTSALYD AEAAVTAAIDTFGKQAILAASRRLTKSAVPKRIRESITDPNAVIASTRAEVMRQTGADQIKAETITRFSR GQLIQLVLIGALVYVAYPFISTVPTFFSQLRTANWWWALLGLAVSALTYVGAAAALWACADGLVGFWKLS IMQVANTFAATTTPAGVGGLALSTRFLQKGGLTAVRATAAVALQQSVQVIVHLVLLILFSALAGTSTDLS HFVPNATVLYLIAGVALGIVGTFLFVPKLRRWLATAVRPKLREVTNDLIALAREPKRLALIVLGCAGTTL GAALALWASIEAFGGGTTFVTVTVVTMVGGTLASAAPTPGGVGAVEAALIGGLAAFGVPAALGVPSVLLY RLLTCWLPVFAGWQVMHWLTRHEMI
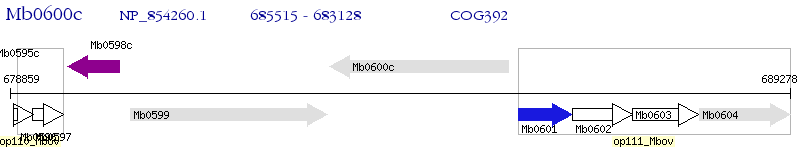
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0600c | - | - | 100% (795) | integral membrane protein |
| M. bovis AF2122 / 97 | Mb0210c | - | 3e-09 | 25.75% (334) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5132 | - | 0.0 | 73.35% (803) | hypothetical protein Mflv_5132 |
| M. tuberculosis H37Rv | Rv0585c | - | 0.0 | 99.87% (795) | integral membrane protein |
| M. leprae Br4923 | MLBr_02618 | - | 3e-09 | 24.09% (328) | putative integral membrane protein |
| M. abscessus ATCC 19977 | MAB_4524 | - | 6e-08 | 23.62% (326) | hypothetical protein MAB_4524 |
| M. marinum M | MMAR_0953 | - | 0.0 | 81.64% (795) | integral membrane protein |
| M. avium 104 | MAV_4559 | - | 0.0 | 78.49% (795) | hypothetical protein MAV_4559 |
| M. smegmatis MC2 155 | MSMEG_1199 | - | 0.0 | 75.09% (795) | hypothetical protein MSMEG_1199 |
| M. thermoresistible (build 8) | TH_0211 | - | 0.0 | 72.69% (487) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_0704 | - | 0.0 | 81.26% (795) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1204 | - | 0.0 | 75.47% (795) | hypothetical protein Mvan_1204 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0600c|M.bovis_AF2122/97 MRVDGRDIGVSGNLLQPLTRRTNDIIRAVLAAIYLVAVITSSLITRPQWV
Rv0585c|M.tuberculosis_H37Rv MRVDGRDIGVSGNLLQPLTRRTNDIIRAVLAAIYLVAVITSSLITRPQWV
MMAR_0953|M.marinum_M MRVDGREITVSGSLLPPITRRTNDIIWLVVAVVFLVVVVTSSVITRPRWV
MUL_0704|M.ulcerans_Agy99 MRVDGREITVSGSLLPPITRRTNDIIWLVVAVVFLVVVVTSSVITRPRWV
MAV_4559|M.avium_104 MRVDGRDIIVSGSLLQPLTRRTNDILRLGMALIFLAVVITGSVITRPQWI
MSMEG_1199|M.smegmatis_MC2_155 MRVDGRDIVVTGKLLQPMSRRTNDILRLTLAALFLATVVASSLITRYEWV
TH_0211|M.thermoresistible__bu --------------------------------------------------
Mflv_5132|M.gilvum_PYR-GCK MRVDGQDVAVSGSLLQPLTRRTNDIFRLSLAGIFLVVVVTSSLITRYEWE
Mvan_1204|M.vanbaalenii_PYR-1 MRVDGQDVAVSGSLLQPLTRRTNDILRLALAGVFLVVIVTSSLITRYEWE
MLBr_02618|M.leprae_Br4923 --------------------------------------------------
MAB_4524|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0600c|M.bovis_AF2122/97 ALEKSISEIVGVLSPSQSDLVYLGYGLAILALPFVILIGLIVSRQWKLLG
Rv0585c|M.tuberculosis_H37Rv ALEKSISEIVGVLSPSQSDLVYLGYGLAILALPFVILIGLIVSRQWKLLG
MMAR_0953|M.marinum_M ALEKSISEIVGVLTPTQSDLVYLAYGAAILALPFVILIGLIVSRQWKLLG
MUL_0704|M.ulcerans_Agy99 ALEKSISEIVGVLTPTQSDLVYLAYGAAILALPFVILIGLIVSRQWKLLG
MAV_4559|M.avium_104 ALEKSVSQIVGVLSPTQSDVVYLVYGLAIVALPFMILIGLIAAGQWKLLG
MSMEG_1199|M.smegmatis_MC2_155 ALERSISEVVGVLTPTQSNLVYLAYSVAIVALPFVILVSLIFSRQWKLLG
TH_0211|M.thermoresistible__bu --------------------------------------------------
Mflv_5132|M.gilvum_PYR-GCK ALEKSISGIVGVLSPTQSNTVYLIYGIAILGLPFFILIGLILARQWKLLG
Mvan_1204|M.vanbaalenii_PYR-1 ALERSISEIVGVLSPTQSNTVYLIYGVAILALPFVILIGLIVSRQWKLLG
MLBr_02618|M.leprae_Br4923 --------------------------------------------------
MAB_4524|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0600c|M.bovis_AF2122/97 AYAAAGLMAVLPLSISSSRIAAPRWHFDLSDRLATLLAQFLDDPRWIAIL
Rv0585c|M.tuberculosis_H37Rv AYAAAGLMAVLPLSISSSRIAAPRWHFDLSDRLATLLAQFLDDPRWIAML
MMAR_0953|M.marinum_M AYAAAAIMAVLPLSISSNRISAPRWHFDLSEKLTTLPAQFLDDPRWIAMV
MUL_0704|M.ulcerans_Agy99 AYAAAAIMAVLPLSISSNRISAPRWHFDLSEKLTTLPAQFLDDPRWIAMV
MAV_4559|M.avium_104 AYAAAGLSAIVLLSISGTGLAAPRWHFDVLDRLTTLPAQLLDDPRWIGML
MSMEG_1199|M.smegmatis_MC2_155 AYAAAGLITMLSLSITGNGIAAPQWHFDLGDRLDSQLSQFLDDPRWIGML
TH_0211|M.thermoresistible__bu --------------------------------------------------
Mflv_5132|M.gilvum_PYR-GCK AYGAAGVIAILALSITGNGIAAPRWHFDLTVRLDTVLSQFLDDPRWIAML
Mvan_1204|M.vanbaalenii_PYR-1 AYAAAGLIAILSLSITGNGIAAPRWHFDLTERLDTVLSQFLDDPRWIAML
MLBr_02618|M.leprae_Br4923 --------------------------------------------------
MAB_4524|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0600c|M.bovis_AF2122/97 AAVLTVSGPWLPARWRHWWWALLLAFVPIHLVVSAIVPARSLLGLAVGWL
Rv0585c|M.tuberculosis_H37Rv AAVLTVSGPWLPARWRHWWWALLLAFVPIHLVVSAIVPARSLLGLAVGWL
MMAR_0953|M.marinum_M AAVLTVSGPWLPARWRHWWWGLLLAFVPIHLVISAIVPARSLLGLAVGWF
MUL_0704|M.ulcerans_Agy99 AAVLTVSGPWLPARWRHWWWGLLLAFVPTHLVISAIVPAGSLLGLAVGWF
MAV_4559|M.avium_104 AAVLTVSGPWLPARWRHWWWALLLAFVPIHLVVSAIVPARSLVGLAVGWV
MSMEG_1199|M.smegmatis_MC2_155 AAVLTVSGPWLPQRLRRWWWGLLLAFVPIHLVVSAVVPARSLLGLAVGWF
TH_0211|M.thermoresistible__bu --------------------------------------------------
Mflv_5132|M.gilvum_PYR-GCK AAVLTVSGPWLPARWRRWWWALLLAFVPIHLVVSAVVPARSLLGLAVGWF
Mvan_1204|M.vanbaalenii_PYR-1 AAVLTVSGPWLPARWRHWWWALLLAFVPIHLVVSAVVPARSLLGLAVGWF
MLBr_02618|M.leprae_Br4923 --------------------------------------------------
MAB_4524|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0600c|M.bovis_AF2122/97 VGALVVLVVGTPALEVPLDGAIRALAKRGFAVSGLAVVRPAGPGPLVLSA
Rv0585c|M.tuberculosis_H37Rv VGALVVLVVGTPALEVPLDGAIRALAKRGFAVSGLAVVRPAGPGPLVLSA
MMAR_0953|M.marinum_M VGALVVLVVGTPALEVPLEGAVRAMAKRGFAVSMLTVVRPGGAGPLVLAA
MUL_0704|M.ulcerans_Agy99 VGALVVLAVGTPALEVPLEGAVRAMAKRGFAVSMLTVVRPGGAGPLVLAA
MAV_4559|M.avium_104 VGALVVLVVGTPALEVPLDAAVRALAKAGFEVCRLTVVRPAGRGPLILSA
MSMEG_1199|M.smegmatis_MC2_155 VGALVVWVVGTPALEVPLDGAVRALMRRGFQVQSFTVVRPAGAGPLLLEA
TH_0211|M.thermoresistible__bu --------------------------------------------------
Mflv_5132|M.gilvum_PYR-GCK VGALVVLVSGTPALEVPLDGAVRALMRRGFPPAELRVLRPAGEGPLVLDA
Mvan_1204|M.vanbaalenii_PYR-1 VGALVVLVVGTPALEVPLDGAVRAMARRGFQVRALKVVRPAGQGPLMLDA
MLBr_02618|M.leprae_Br4923 --------------------------------------------------
MAB_4524|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0600c|M.bovis_AF2122/97 ACEQPNAG--------ACSEALIELYGPHQSGGGALRQLWLKLTLRGTET
Rv0585c|M.tuberculosis_H37Rv ACEQPNAG--------ACSEALIELYGPHQSGGGALRQLWLKLTLRGTET
MMAR_0953|M.marinum_M TSPDPEC------------RAAIELYGPHQRSGGALRQLWWKLRLRGSET
MUL_0704|M.ulcerans_Agy99 TSPDPEC------------RAAIELYGPHQRSGGALRQLWWKLRLRGSET
MAV_4559|M.avium_104 DGQDADH------------TALIELYGPHQRSGGALRQLWGKLKLRDAET
MSMEG_1199|M.smegmatis_MC2_155 ASQDP------------DAVAVVELYGPNQRSGGAFRQLWRKIRFRSDET
TH_0211|M.thermoresistible__bu --------------------------------------------------
Mflv_5132|M.gilvum_PYR-GCK TASCNSKADSKADSDTEPETALVELYGPHQRGVGFLRQFWGKLRLRDDET
Mvan_1204|M.vanbaalenii_PYR-1 TERTS--------DDAEPGTAIVELYGPHQRGGGFLRQFWSKLRLRDSET
MLBr_02618|M.leprae_Br4923 --------------------------------------------------
MAB_4524|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0600c|M.bovis_AF2122/97 APLQASMRRAVEHRALMAIAFGDLGMANTTVIAVSPLDRGWTLYAHRPAR
Rv0585c|M.tuberculosis_H37Rv APLQASMRRAVEHRALMAIAFGDLGMANTTVIAVSPLDRGWTLYAHRPAR
MMAR_0953|M.marinum_M APLQASMRRAVEHRALMAIAIGEAGVANSSTIALAALDREWTLYAHNPVR
MUL_0704|M.ulcerans_Agy99 APLQASMRRAVEHRALMAIAIGEAGVANTSTIALAALDREWTLYAHNPVR
MAV_4559|M.avium_104 APLLTSMRRAVEHRALMAIAIGEAGLANTATVAVATLDRGWMLYSHKPPR
MSMEG_1199|M.smegmatis_MC2_155 APLQTSMRRAVEHRALMAIAIGDLGLASTSTMSVAALDRGWTLYAHNRKR
TH_0211|M.thermoresistible__bu ----------------MAIAADAAGISNTSTLAMSALARGWTMYAHTPAV
Mflv_5132|M.gilvum_PYR-GCK APLQPSMRRAVEHRALMALAVGNLGMANTQPIAVATLDRGWTVYAHKPAR
Mvan_1204|M.vanbaalenii_PYR-1 APIQTSMRRAVEHRALMALAVGDLGMANTSPIAVATLDRGWTVYAHKPAC
MLBr_02618|M.leprae_Br4923 -------------------------MSPAG--------GGEAPP------
MAB_4524|M.abscessus_ATCC_1997 -------------------------MPPSDDRDPDRLEGGCTPPDPNGPD
:. :
Mb0600c|M.bovis_AF2122/97 GIGISECTKTTPTAHVWEALRTLHDQQISHGDLCSAEITVDNGAVLFGGF
Rv0585c|M.tuberculosis_H37Rv GIGISECTKTTPTAHVWEALRTLHDQQISHGDLCSAEITVDNGAVLFGGF
MMAR_0953|M.marinum_M GTPLDECTAQTPVGRVWESLRTLNDYQISHGDLRCHEITVDEDKVLFGGF
MUL_0704|M.ulcerans_Agy99 GTPLDECTAQTPVGRVWESLRTLNDYQISHGDLRCHEITVDEDKVLFGGF
MAV_4559|M.avium_104 GTPIDRCAKTTPVQRLWEALRVLNDHQIAHGDLRAHHITVDDGAVLFGGF
MSMEG_1199|M.smegmatis_MC2_155 GTPLDQCTDEATVGRVWNALRMLHDHQIAHGDLRSKEITVEDGAVLFGGF
TH_0211|M.thermoresistible__bu GVPLAECASTVSVDRVWEALGVCHHHQIAHGDLRPKEITVDNGRVLFGGF
Mflv_5132|M.gilvum_PYR-GCK GIPLADCADETPVARVWDSLGVLHSQQISHGDLRRGEITVVDGRPLFGGF
Mvan_1204|M.vanbaalenii_PYR-1 GTPLDDCADAIPVARVWDSLAVLHSQQISHGDLRGKAITVVDGTPLFGGF
MLBr_02618|M.leprae_Br4923 -----------------------------------------------GKY
MAB_4524|M.abscessus_ATCC_1997 SCPDDPEADTAPQPVIKR----------------------------RGKY
* :
Mb0600c|M.bovis_AF2122/97 GEAEYGATDAQLQSDLAQLLVTTSALYDAEAAVTAAIDTFGKQAILAASR
Rv0585c|M.tuberculosis_H37Rv GEAEYGATDAQLQSDLAQLLVTTSALYDAEAAVTAAIDTFGKQAILAASR
MMAR_0953|M.marinum_M GNAEYGATDAQLQSDIAQLLVTTSALYGAQPAVSAAIEAFGKDTILMASR
MUL_0704|M.ulcerans_Agy99 GNAEYGATDAQLQSDIAQLLVTTSALYGAQPAVSAAIEAFGKDTILMASR
MAV_4559|M.avium_104 GSAEYGATEAQLQSDIAQLLVTTSAHYDPKSAVGAAIDVFGADTILSASR
MSMEG_1199|M.smegmatis_MC2_155 GNSEYGATDAQLQSDIAQLLVTTTALYDAQSAVGAAIKAFGRDTVLTASR
TH_0211|M.thermoresistible__bu SLAEYGATDVQIQADIAQLLVTTTAIYGAEAAVNAAIAAVGKDTVLTASR
Mflv_5132|M.gilvum_PYR-GCK SRAEFGASDAHLHTDIAQLLVTTADLYGPPKAVAAAITVFGNDSVLAASR
Mvan_1204|M.vanbaalenii_PYR-1 AHAEYGASDAHLHTDIAQLLMTTTDLYGAPKAVAAAITVFGFEPVLAASR
MLBr_02618|M.leprae_Br4923 WWVRWVVLSLVAIVLVVELALGWDQLAKAWTSVYEANWWWLLAAVLAAAA
MAB_4524|M.abscessus_ATCC_1997 WWIRWAILIVLAIVLAVEIGFVSDQLDRAWRSLKTANPWWVLAAALAAMA
.: .:: . . :: * . * *
Mb0600c|M.bovis_AF2122/97 RLTKSAVPKRIRESITDPNAVIASTRAEVMRQTGADQIKAETITRFSRGQ
Rv0585c|M.tuberculosis_H37Rv RLTKSAVPKRIRESITDPNAVIASTRAEVMRQTGADQIKAETITRFSRGQ
MMAR_0953|M.marinum_M RLTKSAVPKRIRESVANAKTVIAGARTEVKRQTGADQIESQTITRFSRSQ
MUL_0704|M.ulcerans_Agy99 RLTKSAVPKRIRESVANAKTVIAGARTEVKRQTGADQIESQTITRFSRSQ
MAV_4559|M.avium_104 RLTKVAVPKSVRRTVPDSGAVISGARAEVKRQTGADQIKPQTITRFTRSQ
MSMEG_1199|M.smegmatis_MC2_155 RLTKAAVPARVRATVPDAKAVMGQARDEVKHQTGADQIEAETITRFTRTQ
TH_0211|M.thermoresistible__bu RLTKSAVPARIRKSIDNAGEVIAAARDEVKRQTGADQIDVETITRFNRRQ
Mflv_5132|M.gilvum_PYR-GCK RLTRAAVPKAVRDSVRDAGATMSKLRDEVKYQTGADQIKTETITRFTRNQ
Mvan_1204|M.vanbaalenii_PYR-1 RLTKTAVPKRVRMSVRDAGTVMPTVRDEVKRQTGADQIKTTTITRFTRNQ
MLBr_02618|M.leprae_Br4923 AVHS-----------------FAQIQRTLLKSAGVNVKQLRSEAAFYAAN
MAB_4524|M.abscessus_ATCC_1997 SMHS-----------------FAQIQRTLLRSAGVKIRQWRSEAAFYAGN
: : : : .:*.. . : : * :
Mb0600c|M.bovis_AF2122/97 LIQLVLIGALVYVAYPFISTVPTFFSQLRTANWWWALLGLAVSALTYVGA
Rv0585c|M.tuberculosis_H37Rv LIQLVLIGALVYVAYPFISTVPTFFSQLRTANWWWALLGLAVSALTYVGA
MMAR_0953|M.marinum_M FIQLVLIGALVYVAYPFISTVPTFFSQLRTANWTWALLGLAVSALTYVGA
MUL_0704|M.ulcerans_Agy99 FIQLVLIGALVYVAYPFISTVPTFFSQLRTANWTWALLGLAVSALTYVGA
MAV_4559|M.avium_104 IIQLVLFGALVYVAYPFISTAPTFFSQLRTADWWWALLGLLVSALTYVGA
MSMEG_1199|M.smegmatis_MC2_155 VIQLVLLIALVYVAYPFISTVPTFFSELRTANWSWALMGLTVSALTYVGA
TH_0211|M.thermoresistible__bu VIQLVLLVALVYVAYPFISTVPTFFSELRSVNWWWALAGLAVSASKYVGA
Mflv_5132|M.gilvum_PYR-GCK VVQMVLLVALVYVAYPFISSAPVFFSELRSANWWWALVGLAVSALKYLGA
Mvan_1204|M.vanbaalenii_PYR-1 LIQMVLLVALVYVAYPFISSVPVFFSELRSANWWWALAGLAVSALKYLGA
MLBr_02618|M.leprae_Br4923 SLSTTLPGGPVLSATFLLRQQRIWGASTVVASWQLVMS------------
MAB_4524|M.abscessus_ATCC_1997 ALSTTLPGGPVLSATFIYRQQRLWGATPVVASWQLVMS------------
:. .* . * * : : : ..* .:
Mb0600c|M.bovis_AF2122/97 AAALWACADGLVGFWKLSIMQVANTFAATTTPAGVGGLALSTRFLQKGGL
Rv0585c|M.tuberculosis_H37Rv AAALWACADGLVGFWKLSIMQVANTFAATTTPAGVGGLALSTRFLQKGGL
MMAR_0953|M.marinum_M AAALWTCADGQVSFWKLSIVQVANTFAATTTPAGVGGLALSTRFLQKSGL
MUL_0704|M.ulcerans_Agy99 AAALWTCADGQVSFWKLSIVQVANTFAATTTPAGVGGLALSTRFLQKSGL
MAV_4559|M.avium_104 AAALWACTDGMVNFWMLSIAQVANTFAATTTPAGVGGLALSTRFLQKSGL
MSMEG_1199|M.smegmatis_MC2_155 AAALWACADGLVSFRNLSIMQIANTFAATTTPAGVGGLALSTRFLQKGGL
TH_0211|M.thermoresistible__bu AAALWACANGMVSFRNLTIMQVANTFAATTTPAGVGGLALSTRFLQKGGL
Mflv_5132|M.gilvum_PYR-GCK AAALWACADGMVRFRSLTVMQVANTFAATTTPAGVGGLALSTRFLQKGGL
Mvan_1204|M.vanbaalenii_PYR-1 AAALWACADRLVSFRDLTIMQVANTFAATTTPAGVGGLALSTRFLQKGGL
MLBr_02618|M.leprae_Br4923 ---------GVLQAVGLALLALGGAFFLGAKNN-----PFSLLFALAGFV
MAB_4524|M.abscessus_ATCC_1997 ---------GVLQAVGLAILGLGGAFLLGAKTN-----PFSLVFSVGGLL
: *:: :..:* :. .:* * . :
Mb0600c|M.bovis_AF2122/97 TAVRATAAVALQQSVQVIVHLVLLILFSALAGTSTDLSHFVPNATVLYLI
Rv0585c|M.tuberculosis_H37Rv TAVRATAAVALQQSVQVIVHLVLLILFSALAGTSTDLSHFVPNATVLYLI
MMAR_0953|M.marinum_M SALRATAAVALQQSVQVIVHLALLILFSTVAGTSTDLSHFVPTGTVVYLI
MUL_0704|M.ulcerans_Agy99 SALRATAAVALQQSVQVIVHLALLILFSTVGGTSTDLSHFVPTGTVVYLI
MAV_4559|M.avium_104 SAMRATAAVALQQSVQVIAHLALLVLFSAAAGASMNLSHFVPSATMLYLI
MSMEG_1199|M.smegmatis_MC2_155 STMRATAAVALQQSVQVIVHVILLIFFSTMAGARADLSHFVPSATILYLI
TH_0211|M.thermoresistible__bu GAVRATAAVALQQSVQLTTHIVLLIVFSVAAGTSADLSQFVPDATILYLV
Mflv_5132|M.gilvum_PYR-GCK GALRATTAVALQQSVQVITHITLLILFSAAAGASANLSRFVPSTTVLYLI
Mvan_1204|M.vanbaalenii_PYR-1 GALRATTAVALQQSVQVITHITLLIFFSAAVGVSADLSRFVPSATVLYLI
MLBr_02618|M.leprae_Br4923 TLLLLAQAVASRPELIEGIGRRVLSWVNSVRGKSADTG------------
MAB_4524|M.abscessus_ATCC_1997 AILLLAQAVASRPEMLDGIGVRVLRWVNDLRGKPPMMG------------
: : *** : .: :* .. * .
Mb0600c|M.bovis_AF2122/97 AGVALGIVGTFLFVPKLRRWLATAVRPKLREVTNDLIALAREPKRLALIV
Rv0585c|M.tuberculosis_H37Rv AGVALGIVGTFLFVPKLRRWLATAVRPKLREVTNDLIALAREPKRLALIV
MMAR_0953|M.marinum_M AGVALGIIGTFLFVPNLRRWLSTEVRPKLKEVTSDLIVLGREPKRLAIIL
MUL_0704|M.ulcerans_Agy99 AGVALGIIGTFLFVPNLRRWLSTEVRPKLKEVTSDLIVLGREPKRLAIIL
MAV_4559|M.avium_104 AGVALGIVGTFLFVPTLRRWLATEVRPKLDEVVSDLAKLAREPRRLALIL
MSMEG_1199|M.smegmatis_MC2_155 AGLALGLVGAFLAVPKLRRWLASAVRPKLEEVWEDLIKLAREPKRLILIV
TH_0211|M.thermoresistible__bu VGAALGVIGIAMLVPRLRRWLGTAVRPRLEEVVADLTALAREPARLALIV
Mflv_5132|M.gilvum_PYR-GCK AGVVLGAIGVSLFVPKLRHWLATAVRPRLKEVLDDLMLLAREPKRLALII
Mvan_1204|M.vanbaalenii_PYR-1 AGAALGAVGIFLFVPKLRRWLATAVRPRVKEVANDLIALAREPRRLAVIV
MLBr_02618|M.leprae_Br4923 -------------------------LDKWRQTLTQLESVSLGRRDLAMAF
MAB_4524|M.abscessus_ATCC_1997 -------------------------VARWREILKQMEAVTLTRRALGEAF
: : :: : * .
Mb0600c|M.bovis_AF2122/97 LGCAGTTLGAALALWASIEAFGGGTTFVTVTVVTMVGGTLASAAPTPGGV
Rv0585c|M.tuberculosis_H37Rv LGCAGTTLGAALALWASIEAFGGGTTFVTVTVVTMVGGTLASAAPTPGGV
MMAR_0953|M.marinum_M LGCAGTTLGAALALWASIEAFGGGTTFVTVTVVTMVGGTLASAAPTPGGV
MUL_0704|M.ulcerans_Agy99 LGCAGTTLGAALALWASIEAFGGGTTFVTVTVVTMVGGTLASAAPTPGGV
MAV_4559|M.avium_104 LGCAGTTLGAALALWASIQAFGGDTTFVAVTVVTMVGGTLASAAPTPGGV
MSMEG_1199|M.smegmatis_MC2_155 LGAAGTTLGAALALWASVEAFGGDTSFVTVTVVTMIGGTLASAAPTPGGV
TH_0211|M.thermoresistible__bu AGCGITTLGAALALWASIEAFGGDTSFVTVTIVTMIGGTLASAAPTPGGI
Mflv_5132|M.gilvum_PYR-GCK LGCATTTLGAALALWASIEAFGGDASFITVTIVTMVGGTLASAAPTPGGV
Mvan_1204|M.vanbaalenii_PYR-1 LGCATTTLGGALALWASIEAFGGDASFVTVTIVTMIGGTLASAAPTPGGV
MLBr_02618|M.leprae_Br4923 SWSLFNWIADVACLGFAAYAAGDHASVAGLTVAYAAARAVGTIPLMPGGL
MAB_4524|M.abscessus_ATCC_1997 GWSLFNWIADVACLAFAAYAVGSRPSLVGVMVAYAAARAAGSVPLMPGGL
. . :. . .* : * *. .:. : :. . : .: . ***:
Mb0600c|M.bovis_AF2122/97 GAVEAALIGGLAAFGVPAALGVPSVLLYRLLTCWLPVFAGWQVMHWLTRH
Rv0585c|M.tuberculosis_H37Rv GAVEAALIGGLAAFGVPAALGVPSVLLYRLLTCWLPVFAGWQVMHWLTRH
MMAR_0953|M.marinum_M GAVEAALIGGLAAFGVPAAIGVPSVLLYRVLTCWLPVFLGWPIMRWLTKS
MUL_0704|M.ulcerans_Agy99 GAVEAALIGGLAAFGVPAAIGVPSVLLYRVLTCWLPVFLGWPIMRWLTKS
MAV_4559|M.avium_104 GAVEAALIGGLAAFGVPAAIGVPAVLLYRMLTCWLPVFVGWPVMRWLTKH
MSMEG_1199|M.smegmatis_MC2_155 GAVEAALIGGLAAFGVPPAIGVPAVLLYRVLTCWLPVFVGWPVMRWLTKN
TH_0211|M.thermoresistible__bu GAVEAALIGGLAAFGVPAAIAVPAVLLYRVLTVWIPVFAGWPIMRWLTDN
Mflv_5132|M.gilvum_PYR-GCK GAVEAALIGGLAAFGVPAAVAVPAVLLYRILTCWLPVFVGWPIMRWLTRK
Mvan_1204|M.vanbaalenii_PYR-1 GAVEAALIGGLAAFGVPAAVAVPAVLLYRILTCWLPVFLGWPIMRWLTRK
MLBr_02618|M.leprae_Br4923 LVVEAVLVPGLVSSGMSLPSAISAMLIYRLISWLLIAAIGWVVFFFMFRT
MAB_4524|M.abscessus_ATCC_1997 LVVEAFLVPGLVSSGMTLPDAISAMLIYRAVSWVFISAIGWVIFFFLFRD
.*** *: **.: *:. . .:.::*:** :: : ** :: ::
Mb0600c|M.bovis_AF2122/97 EMI---------------------------------
Rv0585c|M.tuberculosis_H37Rv EMI---------------------------------
MMAR_0953|M.marinum_M EMI---------------------------------
MUL_0704|M.ulcerans_Agy99 EMI---------------------------------
MAV_4559|M.avium_104 EMV---------------------------------
MSMEG_1199|M.smegmatis_MC2_155 DMI---------------------------------
TH_0211|M.thermoresistible__bu DMV---------------------------------
Mflv_5132|M.gilvum_PYR-GCK EMV---------------------------------
Mvan_1204|M.vanbaalenii_PYR-1 DMI---------------------------------
MLBr_02618|M.leprae_Br4923 ENVTDPDD-----------LGCNPDQPLD-------
MAB_4524|M.abscessus_ATCC_1997 ESQIDPDATPAAAKDPDATPGAPPANPPKNSSDPPK
: