For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTFESVADLAAAAGEKVGQSDWVTITQEEVNLFADATGDHQWIHVDPERAAAGPFGTTIAHGFMTLALL PRLQHQMYTVKGVKLAINYGLNKVRFPAPVPVGSRVRATSSLVGVEDLGNGTVQATVSTTVEVEGSAKPA CVAESIVRYVA
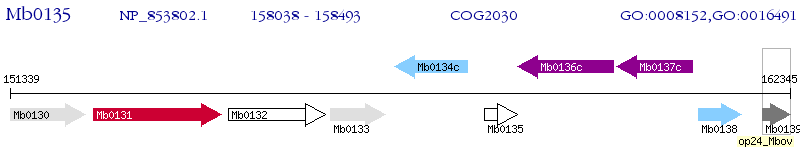
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0135 | - | - | 100% (151) | hypothetical protein Mb0135 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4374 | - | 7e-50 | 62.00% (150) | dehydratase |
| M. tuberculosis H37Rv | Rv0130 | - | 3e-83 | 100.00% (151) | hypothetical protein Rv0130 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1506c | - | 7e-38 | 49.65% (141) | hypothetical protein MAB_1506c |
| M. marinum M | MMAR_0330 | - | 1e-77 | 91.33% (150) | acyl dehydratase |
| M. avium 104 | MAV_5174 | - | 3e-77 | 90.07% (151) | ZbpA protein |
| M. smegmatis MC2 155 | MSMEG_2201 | - | 3e-48 | 58.00% (150) | ZbpA protein |
| M. thermoresistible (build 8) | TH_0017 | - | 1e-50 | 62.67% (150) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4791 | - | 3e-76 | 90.00% (150) | acyl dehydratase |
| M. vanbaalenii PYR-1 | Mvan_1970 | - | 1e-48 | 60.67% (150) | dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4374|M.gilvum_PYR-GCK ------------------MKRFDNLAELVAAQGTQLGPTEWLEITQERVN
Mvan_1970|M.vanbaalenii_PYR-1 MPNIQRRPSSSAGSKETPVKKFNDLAEFVAAEGTQLGPTEWLEITQDRVN
MSMEG_2201|M.smegmatis_MC2_155 ------------------MKVFDNLDALAAAAGTELGPTDWLEVTQDRVN
TH_0017|M.thermoresistible__bu ------------------MKVFNGIEELAAAEGTQLGPTDWLEITQERVN
Mb0135|M.bovis_AF2122/97 ------------------MRTFESVADLAAAAGEKVGQSDWVTITQEEVN
Rv0130|M.tuberculosis_H37Rv ------------------MRTFESVADLAAAAGEKVGQSDWVTITQEEVN
MMAR_0330|M.marinum_M ------------------MRTFESVADLAAAAGETIGQSDWVTITQEDVN
MUL_4791|M.ulcerans_Agy99 ------------------MRTFESVADLAAAAGETIGQSDWVTITQEDVN
MAV_5174|M.avium_104 ------------------MRTFESVADLAAAAGETLGHSDWVTITQEDVN
MAB_1506c|M.abscessus_ATCC_199 -----------------MSTKLATPQELLSLGRVELGTSSWVQITQEQVN
: : : :* :.*: :**: **
Mflv_4374|M.gilvum_PYR-GCK LFADATDDHQWIHVDPARAADGPFGGTIAHGLLTLSLLPRFTHQLYTVDN
Mvan_1970|M.vanbaalenii_PYR-1 LFADATEDHQWIHVDPERAANGPFGGTIAHGLLTLSLLPYFTHQLYTVDN
MSMEG_2201|M.smegmatis_MC2_155 LFADATEDHQWIHVDPEKAKTGPFGGTIAHGLLTLSLLPHFTHQLYRVDN
TH_0017|M.thermoresistible__bu LFADATGDHQWIHVDPDRAAKGPFGGTIAHGLLTLSLLPYFSQQMYRVDN
Mb0135|M.bovis_AF2122/97 LFADATGDHQWIHVDPERAAAGPFGTTIAHGFMTLALLPRLQHQMYTVKG
Rv0130|M.tuberculosis_H37Rv LFADATGDHQWIHVDPERAAAGPFGTTIAHGFMTLALLPRLQHQMYTVKG
MMAR_0330|M.marinum_M LFADATGDHQWIHVDPERAAKGPFGTTIAHGFMTLSLLPRLQHEMYTVKG
MUL_4791|M.ulcerans_Agy99 LFADATGDHQWIHVDPGRAAKGPFGTTIAHGFMTLSLLPRLQHEMYTVKG
MAV_5174|M.avium_104 LFADATGDHQWIHVDPERAASGPFGTTIAHGFMTLALLPRLQHQIYTVNG
MAB_1506c|M.abscessus_ATCC_199 EFADATGDRQWIHIDTERAKSGPFGGTIVHGYLTLALAPAFLDEVLEIAS
***** *:****:*. :* **** **.** :**:* * : .:: : .
Mflv_4374|M.gilvum_PYR-GCK VAMAINYGYNKVRFITPVRVGAKLRARAEISAVAQLEN-AVQATLSTTVE
Mvan_1970|M.vanbaalenii_PYR-1 VAMAINYGYNKVRFITPVRVGAKLRARAEITAVAQLDN-AVQATVSTTVE
MSMEG_2201|M.smegmatis_MC2_155 ITMAINYGYNKVRFITPVRVGAKVRARAQISDVAKLDG-AVQATMTVTVE
TH_0017|M.thermoresistible__bu VKMAINYGYNKVRFINPVKVGARLRGRTEITKVDRLDG-AVQATMTTTVE
Mb0135|M.bovis_AF2122/97 VKLAINYGLNKVRFPAPVPVGSRVRATSSLVGVEDLGNGTVQATVSTTVE
Rv0130|M.tuberculosis_H37Rv VKLAINYGLNKVRFPAPVPVGSRVRATSSLVGVEDLGNGTVQATVSTTVE
MMAR_0330|M.marinum_M IKLAINYGLNKVRFPAPVPVGSRVRAQSSLVGVEDVGNGAVQATVATTVE
MUL_4791|M.ulcerans_Agy99 IKLAINYGLNKVRFPAPVPVGSRVRAQSSLVGVEDVGNGAVQATVATTVE
MAV_5174|M.avium_104 IKLAINYGLNKVRFPAPVPVGSRVRAQSSLVGVDDLGNGAVQATVSTTVE
MAB_1506c|M.abscessus_ATCC_199 YDVVLNYGVNKVRFPAPLHVGARVRGHVTLLSAKRRDKGTVEATYGIKYE
:.:*** ***** *: **:::*. : . :*:** . *
Mflv_4374|M.gilvum_PYR-GCK IEDSEKPAAVAESIVRFIG
Mvan_1970|M.vanbaalenii_PYR-1 IQDSDKPAAVAESIVRFIG
MSMEG_2201|M.smegmatis_MC2_155 IDGSEKPAAVAESIIRFIG
TH_0017|M.thermoresistible__bu IEGEEKPAAVAESIVRIIG
Mb0135|M.bovis_AF2122/97 VEGSAKPACVAESIVRYVA
Rv0130|M.tuberculosis_H37Rv VEGSAKPACVAESIVRYVA
MMAR_0330|M.marinum_M VEGSGKPACVAESVVRYVT
MUL_4791|M.ulcerans_Agy99 VEASGKPACVAESVVRYVT
MAV_5174|M.avium_104 IDGSAKPACVAESIVRYIS
MAB_1506c|M.abscessus_ATCC_199 IEGKDRPPCAAESVYLYQ-
:: . :*...***: