For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKEAINATIQRILRTDRGITANQVLVDDLGFDSLKLFQLITELEDEFDIAISFRDAQNIKTVGDVYTSVA VWFPETAKPAPLGKGTA
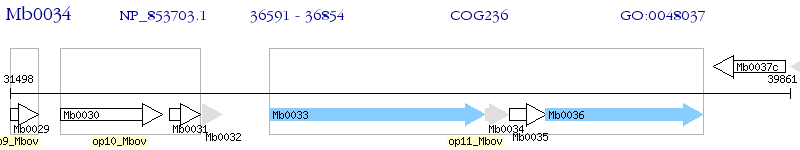
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0034 | acpA | - | 100% (87) | acyl carrier protein AcpA |
| M. bovis AF2122 / 97 | Mb2268 | acpP | 8e-06 | 33.33% (69) | acyl carrier protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4410 | - | 1e-05 | 29.11% (79) | acyl carrier protein |
| M. tuberculosis H37Rv | Rv0033 | acpA | 4e-45 | 100.00% (87) | acyl carrier protein AcpA |
| M. leprae Br4923 | MLBr_01654 | acpP | 8e-07 | 32.88% (73) | acyl carrier protein |
| M. abscessus ATCC 19977 | MAB_1878c | acpP | 9e-06 | 33.33% (69) | acyl carrier protein |
| M. marinum M | MMAR_3337 | acpP | 7e-06 | 32.88% (73) | meromycolate extension acyl carrier protein AcpM |
| M. avium 104 | MAV_2193 | acpP | 7e-06 | 34.78% (69) | acyl carrier protein |
| M. smegmatis MC2 155 | MSMEG_2132 | - | 8e-07 | 33.77% (77) | acyl carrier protein |
| M. thermoresistible (build 8) | TH_1509 | acpM | 1e-05 | 33.33% (69) | MEROMYCOLATE EXTENSION ACYL CARRIER PROTEIN ACPM |
| M. ulcerans Agy99 | MUL_1305 | acpP | 5e-06 | 32.88% (73) | acyl carrier protein |
| M. vanbaalenii PYR-1 | Mvan_3755 | acpP | 3e-05 | 36.17% (47) | acyl carrier protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3337|M.marinum_M -------MAVSQEE--IIAGIAEIIEEVTGIEPSEVTPEKSFVDDLDIDS
MUL_1305|M.ulcerans_Agy99 -------MAVSQEE--IIAGIAEIIEEVTGIEPSEVTPEKSFVDDLDIDS
MAV_2193|M.avium_104 -------MAVSQEE--IIAGIAEIIEEVTGIEPSEVTPEKSFVDDLDIDS
MLBr_01654|M.leprae_Br4923 -------MAVTQEE--IIAGIAEIIEEVTGIEPSEITPEKSFVDDLDIDS
TH_1509|M.thermoresistible__bu -------VATTQEE--IIAGLAEIIEEVTGIEPSEVTPEKSFVDDLDIDS
Mvan_3755|M.vanbaalenii_PYR-1 -------MPATQDE--IIAGLAEIIEEVTGIEPSEVTPEKSFVDDLDIDS
MAB_1878c|M.abscessus_ATCC_199 -------MATAEEKDRIVAGLAEIIEEVTGIEPSEVTLEKSFVDDLDIDS
Mb0034|M.bovis_AF2122/97 ----------MKEA--INATIQRILRTDR-----GITANQVLVDDLGFDS
Rv0033|M.tuberculosis_H37Rv ----------MKEA--INATIQRILRTDR-----GITANQVLVDDLGFDS
Mflv_4410|M.gilvum_PYR-GCK MSTSPAPAAPSPDG--IDAALADILRDDLNVDISRVTRDSRLIDDVGLDS
MSMEG_2132|M.smegmatis_MC2_155 MQST------SPDA--ISAALTEILREDMNVDIRRVTRDSRLVDDVGLDS
: * * : *:. :* :. ::**:.:**
MMAR_3337|M.marinum_M LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVSYIQKLEEENPEAAEAL
MUL_1305|M.ulcerans_Agy99 LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVSYIQKLEEENPEAAEAL
MAV_2193|M.avium_104 LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVAYIQKLEEENPEAAEAL
MLBr_01654|M.leprae_Br4923 LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVTYIQKLEEENPEAAEAL
TH_1509|M.thermoresistible__bu LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVAYIQKLEEENPEAAAAI
Mvan_3755|M.vanbaalenii_PYR-1 LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVAYIQKLEEENPEAAAAI
MAB_1878c|M.abscessus_ATCC_199 LSMVEIAVQTEDKYGVKIPDEDLAGLRTVGDVVNYIEKLEQENPEAAANI
Mb0034|M.bovis_AF2122/97 LKLFQLITELEDEFDIAISFRDAQNIKTVGDVYTSVAVWFPETAKPAPLG
Rv0033|M.tuberculosis_H37Rv LKLFQLITELEDEFDIAISFRDAQNIKTVGDVYTSVAVWFPETAKPAPLG
Mflv_4410|M.gilvum_PYR-GCK VAFAIGMIAIQDKLGVTLNEEDLLSCDTVGDLETAVLAKVPESR------
MSMEG_2132|M.smegmatis_MC2_155 VAFAVGMVAIEDKLGVALSEEDLLSCDTVGDLEAAILAKIPAEQSNQ---
: : :*: .: : .* . ****: :
MMAR_3337|M.marinum_M RAKLETENPEAAANVKARLEAESK
MUL_1305|M.ulcerans_Agy99 RAKLETENPEAAANVKARLEAESK
MAV_2193|M.avium_104 RAKLETENPEAVANVKARLEADSK
MLBr_01654|M.leprae_Br4923 RAKIVSENPEAAANVQARLETESK
TH_1509|M.thermoresistible__bu REKIEAERS---------------
Mvan_3755|M.vanbaalenii_PYR-1 RAQITGDK----------------
MAB_1878c|M.abscessus_ATCC_199 RAQLEGSDK---------------
Mb0034|M.bovis_AF2122/97 KGTA--------------------
Rv0033|M.tuberculosis_H37Rv KGTA--------------------
Mflv_4410|M.gilvum_PYR-GCK ------------------------
MSMEG_2132|M.smegmatis_MC2_155 ------------------------