For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDAATTRVGLTDLTFRLLRESFADAVSWVAKNLPARPAVPVLSGVLLTGSDNGLTISGFDYEVSAEAQVG AEIVSPGSVLVSGRLLSDITRALPNKPVGVHVEGNRVALTCGNARFSLPTMPVEDYPTLPTLPEETGLLP AELFAEAISQVAIAAGRDDTLPMLTGIRVEILGETVVLAATDRFRLAVRELKWSASSPDIEAAVLVPAKT LAEAAKAGIGGSDVRLSLGTGPGVGKDGLLGISGNGKRSTTRLLDAEFPKFRQLLPTEHTAVATMDVAEL IEAIKLVALVADRGAQVRMEFADGSVRLSAGADDVGRAEEDLVVDYAGEPLTIAFNPTYLTDGLSSLRSE RVSFGFTTAGKPALLRPVSGDDRPVAGLNGNGPFPAVSTDYVYLLMPVRLPG
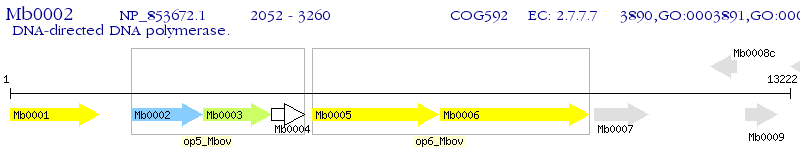
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0002 | dnaN | - | 100% (402) | DNA polymerase III subunit beta |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0826 | - | 1e-174 | 77.61% (402) | DNA polymerase III subunit beta |
| M. tuberculosis H37Rv | Rv0002 | dnaN | 0.0 | 99.75% (402) | DNA polymerase III subunit beta |
| M. leprae Br4923 | MLBr_00002 | dnaN | 1e-178 | 80.85% (402) | DNA polymerase III subunit beta |
| M. abscessus ATCC 19977 | MAB_0002 | dnaN | 1e-159 | 69.40% (402) | DNA polymerase III subunit beta |
| M. marinum M | MMAR_0002 | dnaN | 0.0 | 85.57% (402) | DNA polymerase III (beta chain) DnaN |
| M. avium 104 | MAV_0002 | dnaN | 0.0 | 86.23% (385) | DNA polymerase III subunit beta |
| M. smegmatis MC2 155 | MSMEG_0001 | dnaN | 1e-172 | 77.25% (400) | DNA polymerase III subunit beta |
| M. thermoresistible (build 8) | TH_0848 | dnaN | 1e-174 | 77.44% (399) | DNA POLYMERASE III (BETA CHAIN) DNAN (DNA |
| M. ulcerans Agy99 | MUL_0002 | dnaN | 0.0 | 84.83% (402) | DNA polymerase III subunit beta |
| M. vanbaalenii PYR-1 | Mvan_0002 | - | 1e-177 | 79.35% (397) | DNA polymerase III subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0826|M.gilvum_PYR-GCK -MNVATTAGLSDLKFRLTREDFADAVAWVARNLPSRPTVPVLAGVLLTGS
Mvan_0002|M.vanbaalenii_PYR-1 -MNVPTTAGLTDLKFRLTREDFADAVAWVARNLPSRPTVPVLAGVLLTGS
MSMEG_0001|M.smegmatis_MC2_155 --MATTTAGLTDLKFRVVREDFADAVAWVARSLPTRPTIPVLAGVLLTGT
TH_0848|M.thermoresistible__bu --VATTTVGLTDLKVRLVRDDFADAVAWVARSLPSRPTVPVLAGVLLTGS
Mb0002|M.bovis_AF2122/97 MDAATTRVGLTDLTFRLLRESFADAVSWVAKNLPARPAVPVLSGVLLTGS
Rv0002|M.tuberculosis_H37Rv MDAATTRVGLTDLTFRLLRESFADAVSWVAKNLPARPAVPVLSGVLLTGS
MMAR_0002|M.marinum_M MDAATTRAGLTDLKFRLVRESFADAVSWVAKNLPTRPAVPVLSGVLLTGS
MUL_0002|M.ulcerans_Agy99 MDAATTRAGLTDLKFRLVPESFADAVSWVAKNLPTRPAVPVLSGVLLTGS
MLBr_00002|M.leprae_Br4923 MDLAKTNVGCSDLKFCLARESFASAVSWVAKYLPTRPTVPVLSGVLLTGS
MAV_0002|M.avium_104 -----------------MRESFADAVSWVAKSLPSRPAVPVLSGVLLSGT
MAB_0002|M.abscessus_ATCC_1997 MDLASPTAADTSLKFRVARDDFADSVAWVARSLPSRPTVPVLAGVLLTAH
:.**.:*:***: **:**::***:****:.
Mflv_0826|M.gilvum_PYR-GCK DEGLTVSGFDYEVSAEVQIPAEIASPGSVLVSGRLLSDIVRSLPAKPVDV
Mvan_0002|M.vanbaalenii_PYR-1 DEGLTISGFDYEVSAEVQIPAEIASPGSVLVSGRLLSDIVRALPAKPVDV
MSMEG_0001|M.smegmatis_MC2_155 DEGLTISGFDYEVSAEVKVSAEIASAGSVLVSGRLLSDITKALPAKPVEV
TH_0848|M.thermoresistible__bu DDGLTISSFDYEVSAEVQIPAEIAAPGTVLVSGRLLSEITRALPNKPVDL
Mb0002|M.bovis_AF2122/97 DNGLTISGFDYEVSAEAQVGAEIVSPGSVLVSGRLLSDITRALPNKPVGV
Rv0002|M.tuberculosis_H37Rv DNGLTISGFDYEVSAEAQVGAEIVSPGSVLVSGRLLSDITRALPNKPVDV
MMAR_0002|M.marinum_M DEGLTISGFDYEVSAEVRVPAEIASPGTVLVSGRLLSDITRALPNKPVDF
MUL_0002|M.ulcerans_Agy99 DEGLTISGFDYEVSAEVRVPAEIASPGTVLVSGRLLSDITRALPKKPVDF
MLBr_00002|M.leprae_Br4923 DSGLTISGFDYEVSAEVQVAAEIASSGSVLVSGRLLSDITRALPNKPVHF
MAV_0002|M.avium_104 DEGLTISGFDYEVSAEAQVAAEIASPGSVLVSGRLLSDIVRALPNKPIDF
MAB_0002|M.abscessus_ATCC_1997 DTGLTLSGFDYEVSAQVKVPAEVAAAGNALVSGRLLSDITRSLPNKPVDV
* ***:*.*******:.:: **:.:.*..********:*.::** **: .
Mflv_0826|M.gilvum_PYR-GCK SVEGTRVSLTCGSARFSLPTMAVEDYPTLPTLPDETGVVSADLFAEAIGQ
Mvan_0002|M.vanbaalenii_PYR-1 SVEGTRVSLTCGSARFSLPTMPVEDYPTLPTLPDETGVVSSDLFAEAIGQ
MSMEG_0001|M.smegmatis_MC2_155 SVEGTRVSLTCGSARFSLPTLAVEDYPALPALPEETGVIASDLFAEAIGQ
TH_0848|M.thermoresistible__bu SVEGTRVSLTCGSARFSLPTMAVEDYPALPELPAETGSVPADLFAEAIGQ
Mb0002|M.bovis_AF2122/97 HVEGNRVALTCGNARFSLPTMPVEDYPTLPTLPEETGLLPAELFAEAISQ
Rv0002|M.tuberculosis_H37Rv HVEGNRVALTCGNARFSLPTMPVEDYPTLPTLPEETGLLPAELFAEAISQ
MMAR_0002|M.marinum_M YVDGNRVALTCGNARFSLPTMAVEDYPTLPTLPEDTGTLPAELFSEAIGQ
MUL_0002|M.ulcerans_Agy99 YVDGNRVALTCGNARFSLPTMAVEDYPTLPTLPEDTGTLPAELFSEAIGQ
MLBr_00002|M.leprae_Br4923 YVDGNRVALTCGSARFSLPTMAVEDYPTLPTLPDETGTLPSDVFAEAIGQ
MAV_0002|M.avium_104 YVDGNRVALNCGSARFSLPTMAVEDYPTLPTLPEETGTLPADLFAEAIGQ
MAB_0002|M.abscessus_ATCC_1997 ALEGTRLLISCGSAKFSLPSMPVEDYPALPSLPEETGALGADVFSEAIGQ
::*.*: :.**.*:****::.*****:** ** :** : :::*:***.*
Mflv_0826|M.gilvum_PYR-GCK VAVAAGRDDTLPMLTGIRVEISGEKVVLAATDRFRLAVRELTWSAGAAGV
Mvan_0002|M.vanbaalenii_PYR-1 VAVAAGRDDTLPMLTGIRVEISGEKVVLAATDRFRLAVRELTWSTGAAGV
MSMEG_0001|M.smegmatis_MC2_155 VAVAAGRDDTLPMLTGIRVEISGESVVLAATDRFRLAVRELTWVTTAGDV
TH_0848|M.thermoresistible__bu VAVAAGRDDTLPMLTGIRVEISGDRMVLAATDRFRLAVRELTWTTKTPDV
Mb0002|M.bovis_AF2122/97 VAIAAGRDDTLPMLTGIRVEILGETVVLAATDRFRLAVRELKWSASSPDI
Rv0002|M.tuberculosis_H37Rv VAIAAGRDDTLPMLTGIRVEILGETVVLAATDRFRLAVRELKWSASSPDI
MMAR_0002|M.marinum_M VAIAAGRDDTLPMLTGIRVEISGEKVVLAATDRFRLAVRELTWNALSPDI
MUL_0002|M.ulcerans_Agy99 VAITAGRDDTLPMLTGIRVEISGEKVVLAATDRFRLAVRELTWNALSPDI
MLBr_00002|M.leprae_Br4923 VAIAAGRDYTLPMLTGIRIEISGDTVVLAATDRFRLAVRELKWSVLSSDF
MAV_0002|M.avium_104 VAIAAGRDDTLPMLTGIRVEISGDTVVLAATDRFRLAVRELTWSAASPDI
MAB_0002|M.abscessus_ATCC_1997 VAVAAGKDDTLPMLTGIRVEINGDTVVLAATDRFRLAVRELNWTSGSGET
**::**:* *********:** *: :***************.* :
Mflv_0826|M.gilvum_PYR-GCK EAAVLVPAKTLAEAAKAGTDGTEVHLALGQGPTVGKEGLLGIRSKGKRST
Mvan_0002|M.vanbaalenii_PYR-1 EAAVLVPAKTLAEAAKAGTDGNEVHLALGAGPSVGKEGLLGIRSKGKRST
MSMEG_0001|M.smegmatis_MC2_155 EAAVLVPAKTLAEAAKAGTDGNQVHLALGSGASVGKDGLLGIRSEGKRST
TH_0848|M.thermoresistible__bu EAAVLVPAKTLAEAAKTGLDGSEVQLALGAGPSVGQDGLLGIRSEGKRST
Mb0002|M.bovis_AF2122/97 EAAVLVPAKTLAEAAKAGIGGSDVRLSLGTGPGVGKDGLLGISGNGKRST
Rv0002|M.tuberculosis_H37Rv EAAVLVPAKTLAEAAKAGIGGSDVRLSLGTGPGVGKDGLLGISGNGKRST
MMAR_0002|M.marinum_M EAAVLVPAKTLGEAAKANTDGSEVRLSLGAGMGVGKDGLLGISGDGKRST
MUL_0002|M.ulcerans_Agy99 EAAVLVPAKTLGEAAKANTDGSEVRLSLGAGMGVGKDGLLGISGDGKRST
MLBr_00002|M.leprae_Br4923 EASVLVPAKTLVEVAKAGTDGSGVCLSLGAGVGVGKDGLFGISGGGKRST
MAV_0002|M.avium_104 EAAVLVPAKTLAEAARTGIDGSDVRLSLGAGAGVGKDGLLGISGNGKRST
MAB_0002|M.abscessus_ATCC_1997 SAAVLVPAKTLSEAAKSASSGSDVQLALGAGSAIGSEGLLGIVSDGKRST
.*:******** *.*:: .*. * *:** * :*.:**:** . *****
Mflv_0826|M.gilvum_PYR-GCK TRLLDAEFPKFRQLLPTEHTAIATIGVAELTEAIKRVALVADRGAQVRME
Mvan_0002|M.vanbaalenii_PYR-1 TRLLDAEFPKFRQLLPTEHTAIATIGVAELTEAIKRVALVADRGAQVRME
MSMEG_0001|M.smegmatis_MC2_155 TRLLDAEFPKFRQLLPAEHTAVATIGVAELTEAIKRVALVADRGAQIRME
TH_0848|M.thermoresistible__bu TRLLDAEFPKFRQLLPTEHTAMATIGVGELTEAIKRVALVADRGAQVRME
Mb0002|M.bovis_AF2122/97 TRLLDAEFPKFRQLLPTEHTAVATMDVAELIEAIKLVALVADRGAQVRME
Rv0002|M.tuberculosis_H37Rv TRLLDAEFPKFRQLLPTEHTAVATMDVAELIEAIKLVALVADRGAQVRME
MMAR_0002|M.marinum_M TRLLDAEFPKFRQLLPAEHTAVASADVAELTEAIKLVALVADRGAQVRME
MUL_0002|M.ulcerans_Agy99 TRLLDAEFPKFRQLLPAEHTAVASADVAELTEAIKLVALVADRGAQVRME
MLBr_00002|M.leprae_Br4923 TRLLDAEFPKFRQLLPAEHTAVATIDVAELTEAIKLVALVADRGAQVRME
MAV_0002|M.avium_104 TRLLDAEFPKFRQLLPAEHTAVATINVAELTEAIKLVALVADRGAQVRME
MAB_0002|M.abscessus_ATCC_1997 TRLLDAEFPKFRQLLPTEHTALATIAISELVEAIKRVALVADRGAQIRLE
****************:****:*: :.** **** **********:*:*
Mflv_0826|M.gilvum_PYR-GCK FADDVLRLSAGADDVGRAEEDLPVQFSGDPLTIAFNPSYLTDGLGSLHAE
Mvan_0002|M.vanbaalenii_PYR-1 FADGVLRLSAGADDVGRAEEDLPVEFSGDPLTIAFNPTYLTDGLSSLHAE
MSMEG_0001|M.smegmatis_MC2_155 FSDDTLKLSAGADDVGRAEEDLPVDFAGEPLTIAFNPTYLTDGLGSLHSE
TH_0848|M.thermoresistible__bu FADDVLHLSAGADDVGRAEEDLPVSFSGEPLTIAFNPGYLTDGLGALHSE
Mb0002|M.bovis_AF2122/97 FADGSVRLSAGADDVGRAEEDLVVDYAGEPLTIAFNPTYLTDGLSSLRSE
Rv0002|M.tuberculosis_H37Rv FADGSVRLSAGADDVGRAEEDLVVDYAGEPLTIAFNPTYLTDGLSSLRSE
MMAR_0002|M.marinum_M FADGMLRLSAGADDVGRAEEDIAVEFAGQPLTIAFNPTYLIDGLASLHSE
MUL_0002|M.ulcerans_Agy99 FADGMLRLSAGADDVGRAEEDIAVEFAGQPLTIAFNPTYLIDGLASLHSE
MLBr_00002|M.leprae_Br4923 FGDGILRLSAGADDVGRAEEDLAVAFTGEPLTIAFNPNYLTDGLASVHSE
MAV_0002|M.avium_104 FSEGSLRLSAGADDVGRAEEDLAVDFAGEPLTIAFNPTYLTDGLGSVRSE
MAB_0002|M.abscessus_ATCC_1997 FEPGLLRLSAGADDVGRAEEELEVEFVGEPLTIAFNPTYLTDGLGSLHSN
* . ::*************:: * : *:******** ** ***.:::::
Mflv_0826|M.gilvum_PYR-GCK RVTFGFTTPSRPAVLRPAGDDD---GSAAGSGPYPAAQTDYVYLLMPVRL
Mvan_0002|M.vanbaalenii_PYR-1 RVTFGFTTPSRPAVLRPAAEDD---GGAGGSGPFPAAQTDYVYLLMPVRL
MSMEG_0001|M.smegmatis_MC2_155 RVTFGFTTPSRPAVLRPAGEDD---GANGGSGPFPAAKTDYVYLLMPVRL
TH_0848|M.thermoresistible__bu RVTFGFTTPSKPAVLRPATEAD---AALNGNGPFPAAETDYVYLLMPVRL
Mb0002|M.bovis_AF2122/97 RVSFGFTTAGKPALLRPVSGDDRPVAGLNGNGPFPAVSTDYVYLLMPVRL
Rv0002|M.tuberculosis_H37Rv RVSFGFTTAGKPALLRPVSGDDRPVAGLNGNGPFPAVSTDYVYLLMPVRL
MMAR_0002|M.marinum_M RVSFGFTTPGKPALLRPASAEDRIVSGPGDQGPFAAVPTDYVYLLMPVRL
MUL_0002|M.ulcerans_Agy99 RVSFGFTTPGKPALLRPASAEDRIVSGPGDQGPFAAVPTDYVYLLMPVRL
MLBr_00002|M.leprae_Br4923 RVSFGFTTPSKPALLRPTSNDD---VHPTHDGPFPALPTDYVYLLMPVRL
MAV_0002|M.avium_104 RVSFGFTTPGKPALLRPASDDD---SPPSGSGPFSALPTDYVYLLMPVRL
MAB_0002|M.abscessus_ATCC_1997 RVTFGFTTPSRPAVLRPADDDQ---SSPEGSGPFTAAQTDYVYLLMPVRL
**:*****..:**:***. : .**:.* ************
Mflv_0826|M.gilvum_PYR-GCK PG
Mvan_0002|M.vanbaalenii_PYR-1 PG
MSMEG_0001|M.smegmatis_MC2_155 PG
TH_0848|M.thermoresistible__bu PG
Mb0002|M.bovis_AF2122/97 PG
Rv0002|M.tuberculosis_H37Rv PG
MMAR_0002|M.marinum_M PG
MUL_0002|M.ulcerans_Agy99 PG
MLBr_00002|M.leprae_Br4923 PG
MAV_0002|M.avium_104 PG
MAB_0002|M.abscessus_ATCC_1997 PG
**