For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LMVAVRVASLPGLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIQPGYFADHQPPALLMLHPPS ETTPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGTITSMVSRVGVDPISHPDTAILPMLLAA
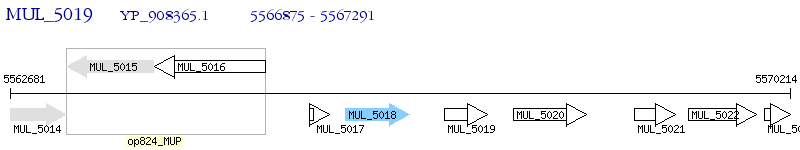
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_5019 | - | - | 100% (138) | hypothetical protein MUL_5019 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3877 | - | 5e-76 | 99.28% (138) | hypothetical protein Mb3877 |
| M. gilvum PYR-GCK | Mflv_1134 | - | 1e-69 | 89.13% (138) | hypothetical protein Mflv_1134 |
| M. tuberculosis H37Rv | Rv3847 | - | 5e-76 | 99.28% (138) | hypothetical protein Rv3847 |
| M. leprae Br4923 | MLBr_00071 | - | 1e-75 | 97.83% (138) | hypothetical protein MLBr_00071 |
| M. abscessus ATCC 19977 | MAB_0117c | - | 8e-64 | 78.99% (138) | hypothetical protein MAB_0117c |
| M. marinum M | MMAR_5397 | - | 7e-76 | 99.28% (138) | hypothetical protein MMAR_5397 |
| M. avium 104 | MAV_0181 | - | 5e-74 | 97.10% (138) | hypothetical protein MAV_0181 |
| M. smegmatis MC2 155 | MSMEG_6428 | - | 1e-68 | 87.68% (138) | hypothetical protein MSMEG_6428 |
| M. thermoresistible (build 8) | TH_0751 | - | 2e-66 | 84.78% (138) | HYPOTHETICAL PROTEIN Rv3847 |
| M. vanbaalenii PYR-1 | Mvan_5672 | - | 7e-70 | 89.13% (138) | hypothetical protein Mvan_5672 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1134|M.gilvum_PYR-GCK MDFGSGRAIEIAPFHSRGSLKGFVVSGRWPDSTKEWAQLLMAAVRVASLP
Mvan_5672|M.vanbaalenii_PYR-1 MDFGSGRAIEIAPFHSRGSLKGFVVSGRWPDSTKEWAQLLMVAVRVASMP
MSMEG_6428|M.smegmatis_MC2_155 MDNRSGRAIEIAPFHSGGSLKGFVVSGRWPDSTKEWAQLLMATVRVASWP
TH_0751|M.thermoresistible__bu MDKGSDRAIEIAPFHLRGALKGFVVSGRWPNSTKEWAQLLKAAVRVASVP
Mb3877|M.bovis_AF2122/97 MGTGSGGPIGVSPFHSRGALKGFVISGRWPDSTKEWAQLLMVAVRVASLP
Rv3847|M.tuberculosis_H37Rv MGTGSGGPIGVSPFHSRGALKGFVISGRWPDSTKEWAQLLMVAVRVASLP
MUL_5019|M.ulcerans_Agy99 ---------------------------------------MMVAVRVASLP
MMAR_5397|M.marinum_M MDAGSDGPIGVAPFHSRGSLKGFVISGRWPDSTKEWAQLLMVAVRVASLP
MLBr_00071|M.leprae_Br4923 METGSGLPIGVVPFHARGALKGFVISGRWPDSTKEWAQLLMVAVRIASLP
MAV_0181|M.avium_104 -----------------------MISGRWPDSTKEWAQLLMVAVRVASLP
MAB_0117c|M.abscessus_ATCC_199 MATSSDQPIEIAPFHAGGSLRGFVVCGRWPDSTKEWMQLLIVTVRIATLP
: .:**:*: *
Mflv_1134|M.gilvum_PYR-GCK GLLSTTTIFGVREELPEQPEPGTVGLVLAEGTVVGESAVPPGHFADRQPP
Mvan_5672|M.vanbaalenii_PYR-1 GLLSTTTIFGAREELPDEPEPGTVGLVLAEGTVVGDAAVPPGHFADRQPP
MSMEG_6428|M.smegmatis_MC2_155 GLLSTTTIFGVREELPDEPQPGTVGLVMAEGPVVGEAAVPPGYFAEHQPP
TH_0751|M.thermoresistible__bu GLLPTTTVFGVREELPDDPEPGTVGLVVAEGPVMGESAVPPGHFADKQPP
Mb3877|M.bovis_AF2122/97 GLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIQPGYFADHQPP
Rv3847|M.tuberculosis_H37Rv GLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIQPGYFADHQPP
MUL_5019|M.ulcerans_Agy99 GLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIQPGYFADHQPP
MMAR_5397|M.marinum_M GLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIQPGYFADHQPP
MLBr_00071|M.leprae_Br4923 GLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIQPGYFADHQPP
MAV_0181|M.avium_104 GLLSTTTVFGAREELPDEPEPGTVGLVLAEGTVFGESAIPPGHFADHQPP
MAB_0117c|M.abscessus_ATCC_199 GLLSTTTIFGAREDLPDDPAPGMVGLVIAEGTVLGESAVAPGRFAEHQPP
***.***:**.**:**::* ** ****:***.*.*::*: ** **::***
Mflv_1134|M.gilvum_PYR-GCK ALMMLHPPSETTPSLPECTGAASGCVLLPGLPHLGLEHRAAWVEAESDGT
Mvan_5672|M.vanbaalenii_PYR-1 ALMMLHPPSETTPSLPECAGAASGCVLLPGLPHLGLEHRAAWVEAESDGT
MSMEG_6428|M.smegmatis_MC2_155 ALLMLHPPSETTPSLPECAGAASGCVLLPGLPHLGLEHRAAWVEAEADGT
TH_0751|M.thermoresistible__bu ALMLLHPPSETTPSLPECAGAASGCVLLPGIPHLGLEHRAAWVEAEADGT
Mb3877|M.bovis_AF2122/97 ALLMLHPPSETTPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGT
Rv3847|M.tuberculosis_H37Rv ALLMLHPPSETTPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGT
MUL_5019|M.ulcerans_Agy99 ALLMLHPPSETTPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGT
MMAR_5397|M.marinum_M ALLMLHPPSETTPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGT
MLBr_00071|M.leprae_Br4923 ALLMLHPPSETMPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGT
MAV_0181|M.avium_104 ALLMLHPPSETMPSLPECTGAASGCVLLPGLPYLGLEHRAAWVEAEADGT
MAB_0117c|M.abscessus_ATCC_199 ALLMLHPPSETNPTLPECLGAASGCLLLPGIPHLGLDHRAAWVEAESDGT
**::******* *:**** ******:****:*:***:*********:***
Mflv_1134|M.gilvum_PYR-GCK VTSMVSRVGVDPISHPDTAILAMLLAA
Mvan_5672|M.vanbaalenii_PYR-1 VTSMVSRVGVDPISHPDTAILAMLLAA
MSMEG_6428|M.smegmatis_MC2_155 VTSMVSRVGVDPISHPDTAILAMLLAA
TH_0751|M.thermoresistible__bu VTSMMSRVGVDPISHPDTAILAMLLAA
Mb3877|M.bovis_AF2122/97 ITSMVSRVGVDPISHPDTAILAMLLAA
Rv3847|M.tuberculosis_H37Rv ITSMVSRVGVDPISHPDTAILAMLLAA
MUL_5019|M.ulcerans_Agy99 ITSMVSRVGVDPISHPDTAILPMLLAA
MMAR_5397|M.marinum_M ITSMVSRVGVDPISHPDTAILAMLLAA
MLBr_00071|M.leprae_Br4923 ITSMVSRVGVDPISHPDTAILAMLLAA
MAV_0181|M.avium_104 ITSMVSRVGVDPISHPDTAILAMLLAA
MAB_0117c|M.abscessus_ATCC_199 VTSVVSRVGIDPISDPDTAVLAMLLAA
:**::****:****.****:*.*****