For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSFPAAPPAVPPVVRRLAAGRPVQAVWANELGGVTFRVGPGAEFIKVARTGVFDFTAEAQRLRWASAHLT VPAVLDFGVEQDWAWLRTAGLPGVSAVHPRWVAAPDVAVRAIGVGLRTLHERLPVRACPFDWSAQSRLAR LTAAGRANVGAPPPIDRLVVCHGDACAPHTLIAEDGNCCGHVDLGTLGVADRWADLAVATLSLQWNFPGH GWEAKFFAAYGVAPDPRRIDYYRRLWGPRISTQAKLDAAFGAEHASAVAVAGRYLCRSQASATPAF
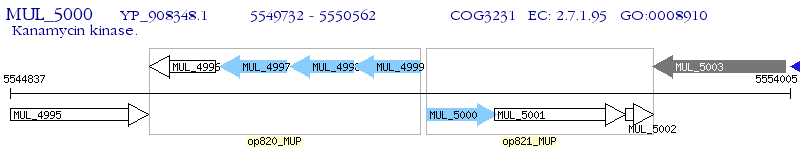
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_5000 | - | - | 100% (276) | phosphotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3847 | - | 8e-98 | 68.31% (243) | putative phosphotransferase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv3817 | - | 8e-98 | 68.31% (243) | phosphotransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0163c | - | 6e-76 | 57.74% (239) | phosphotransferase |
| M. marinum M | MMAR_5380 | - | 1e-141 | 99.58% (237) | phosphotransferase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3847|M.bovis_AF2122/97 MSFPSSPPALPAIVARFAVGRPVRAVWVNELGGVTFRVDSGMGAGCEFIK
Rv3817|M.tuberculosis_H37Rv MSFPSSPPALPAIVARFAVGRPVRAVWVNELGGVTFRVDSGMGAGCEFIK
MUL_5000|M.ulcerans_Agy99 MSFPAAPPAVPPVVRRLAAGRPVQAVWANELGGVTFRVGP----GAEFIK
MMAR_5380|M.marinum_M MSFPAAPPAVPPVVRRLAAGRPVQAVWANELGGVTFRVGP----GAEFIK
MAB_0163c|M.abscessus_ATCC_199 MTIPTEPVTIPDIVAHLSAGRPVLAVWHNQIGGHTFQIGSG--ADREFVK
*::*: * ::* :* :::.**** *** *::** **::.. . **:*
Mb3847|M.bovis_AF2122/97 VARRG-TADFANEARRLRWAAPYLAVPRVLGVGVDGDWAWLHTDALPGLS
Rv3817|M.tuberculosis_H37Rv VARRG-TADFANEARRLRWAAPYLAVPRVLGVGVDGDWAWLHTDALPGLS
MUL_5000|M.ulcerans_Agy99 VARTG-VFDFTAEAQRLRWASAHLTVPAVLDFGVEQDWAWLRTAGLPGVS
MMAR_5380|M.marinum_M VARTG-VFDFTAEAQRLRWASAHLTVPAVLDFGVEQDWAWLRTAGLPGVS
MAB_0163c|M.abscessus_ATCC_199 VSAPHPAINLPAEAERLRWARRYTTVPTVLSVNTEGPLHWLHTAGIPGDS
*: . ::. **.***** : :** **....: **:* .:** *
Mb3847|M.bovis_AF2122/97 AVHPRWRASPQVAVPALGAGLRTLHDSLPVHSCPFDWSTASRLAKLAPAR
Rv3817|M.tuberculosis_H37Rv AVHPRWRASPQVAVPALGAGLRTLHDSLPVHSCPFDWSTASRLAKLAPAR
MUL_5000|M.ulcerans_Agy99 AVHPRWVAAPDVAVRAIGVGLRTLHERLPVRACPFDWSAQSRLARLTAAG
MMAR_5380|M.marinum_M AVHPRWVAAPDVAVRAIGVGLRTLHERLPVRACPFDWSAQSRLARLTAAG
MAB_0163c|M.abscessus_ATCC_199 AVAPRWIENPEVSVRAIASGLRALHDRLPVVDCPYSWSVAARLARIPNPG
** *** *:*:* *:. ***:**: *** **:.**. :***::. .
Mb3847|M.bovis_AF2122/97 RAELGDSPPVDRLVVCHGDACSPNTILDDTGRCCGHVDFGNLGVADRWAD
Rv3817|M.tuberculosis_H37Rv RAELGDSPPVDRLVVCHGDACSPNTILDDTGRCCGHVDFGNLGVADRWAD
MUL_5000|M.ulcerans_Agy99 RANVGAPPPIDRLVVCHGDACAPHTLIAEDGNCCGHVDLGTLGVADRWAD
MMAR_5380|M.marinum_M RANVGAPPPIDRLVVCHGDACAPNTLIAEDGNCCGHVDLGTLGVADRWAD
MAB_0163c|M.abscessus_ATCC_199 --GLSEHPAIDRLVVCHGDACAPNTLLDGNGNWCGHVDFGDLGVADRWAD
:. *.:***********:*:*:: *. *****:* *********
Mb3847|M.bovis_AF2122/97 LAVATLSLQWNFPDYPGQVRDDEFFAAYGVAPDPARIDYYRRLWQ-----
Rv3817|M.tuberculosis_H37Rv LAVATLSLQWNFPDYPGQVRDDEFFAAYGVAPDPARIDYYRRLWQ-----
MUL_5000|M.ulcerans_Agy99 LAVATLSLQWNFP---GHGWEAKFFAAYGVAPDPRRIDYYRRLWGPRIST
MMAR_5380|M.marinum_M LAVATLSLQWNFP---GHGWEAKFFAAYGVAPDPRRIDYYRRLWG-----
MAB_0163c|M.abscessus_ATCC_199 LAVASWSLEWNYG----KGWNDAFFAAYGVTEDPERMAYYRRLWD-----
****: **:**: : : *******: ** *: ******
Mb3847|M.bovis_AF2122/97 -AEDDSSR--------------------------
Rv3817|M.tuberculosis_H37Rv -AEDDSSR--------------------------
MUL_5000|M.ulcerans_Agy99 QAKLDAAFGAEHASAVAVAGRYLCRSQASATPAF
MMAR_5380|M.marinum_M AEDLDSS---------------------------
MAB_0163c|M.abscessus_ATCC_199 -SEE------------------------------
.