For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGEGPMAYHNPFIVNGKIRFPENTNLVRHVEKWARVRGDKLAYRFLDFSTERDGVERDILWSEFSARNRA VGARLQQVTQPGDRIAILCPQNLDYLISFFGALYSGRIAVPLFDPAEPGHVGRLHAVLDDCTPSTILTST DSAEGVRKFIRSRSAKKRPRVIAVDAVPTEVASTWQQPEANELTTAYLQYTSGSTRVPSGVQITHLNLPT NVLQVLNALEGQEGDRGVSWLPFFHDMGLITVLLASVLGHSFTFMTPAAFVRRPGRWIRELARKPGETGG TFSAAPNFAFEHAAMRGVPRDDEPPLDLSNVKGILNGSEPVSPASMRKFFKAFEPYGLRETAVKPSYGLA EATLFVSTTPMDEVPTVIHVDRDELNKQRFVEVAADAPNAVAQVSAGKVGVDEWAVIVDTETASELPDGQ IGEIWLHGNNLGIGYWGKEEESAQTFRNILKSRVPESHAEGAPDDGLWVRTGDYGTYFKGHLYIAGRIKD LVIIDGRNHYPQDLEYTAQESTKALRVGYVAAFSVPANQLPQKVFDDPHAGLSFDPEDTSEQLVIVGERA AGTHKLEYQPIADDIRAAIAVGHGVTVRDVLLVSAGTIPRTSSGKIGRRACRTAYIDGSLRSGVSSPTVF ATGS*
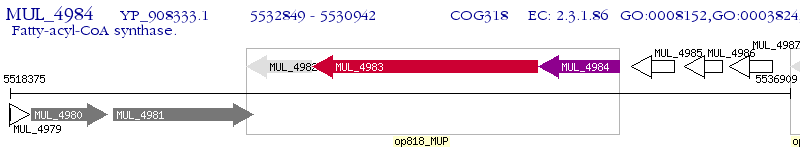
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4984 | fadD32 | - | 100% (635) | acyl-CoA synthetase |
| M. ulcerans Agy99 | MUL_2920 | fadD31 | e-153 | 47.13% (609) | acyl-CoA synthetase |
| M. ulcerans Agy99 | MUL_2020 | fadD26 | e-104 | 39.80% (598) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3831c | fadD32 | 0.0 | 93.36% (633) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_1168 | - | 0.0 | 73.95% (618) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv3801c | fadD32 | 0.0 | 93.21% (633) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_00100 | fadD32 | 0.0 | 90.98% (632) | long-chain-fatty-acid--CoA ligase |
| M. abscessus ATCC 19977 | MAB_0179 | - | 0.0 | 66.77% (626) | acyl-CoA synthetase |
| M. marinum M | MMAR_5365 | fadD32 | 0.0 | 99.68% (629) | fatty acyl-AMP ligase FadD32 |
| M. avium 104 | MAV_0217 | - | 0.0 | 84.31% (631) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_6393 | - | 0.0 | 74.80% (619) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_1609 | - | 0.0 | 75.16% (479) | acyl-CoA synthase |
| M. vanbaalenii PYR-1 | Mvan_5641 | - | 0.0 | 75.40% (618) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1168|M.gilvum_PYR-GCK --------MAFHNPFLK-DGLIKFPDNGNLVRHVERWAKVRGDKMAYRFI
Mvan_5641|M.vanbaalenii_PYR-1 --------MAFHNPFIK-DGLIKFPDNGSLVKHVERWAKVRGDKVAYRFL
MSMEG_6393|M.smegmatis_MC2_155 --------MPFHNPFIK-DGQIKFPDGSSIVAHVERWAKVRGDKLAYRFL
TH_1609|M.thermoresistible__bu --------------------------------------------------
MUL_4984|M.ulcerans_Agy99 --MTGEGPMAYHNPFIV-NGKIRFPENTNLVRHVEKWARVRGDKLAYRFL
MMAR_5365|M.marinum_M --------MAYHNPFIV-NGKIRFPENTNLVRHVEKWARVRGDKLAYRFL
Mb3831c|M.bovis_AF2122/97 MFVTGESGMAYHNPFIV-NGKIRFPANTNLVRHVEKWAKVRGDKLAYRFL
Rv3801c|M.tuberculosis_H37Rv MFVTGESGMAYHNPFIV-NGKIRFPANTNLVRHVEKWAKVRGDKLAYRFL
MLBr_00100|M.leprae_Br4923 --MTGEGRMAYHNPFIV-NGKIRFPPNTNLVRHVEKWAKVRGDKLAYRFL
MAV_0217|M.avium_104 --------MAYHNPFIV-NGKIKFPENTNLVKHVEKWARVRGDKLAYRFL
MAB_0179|M.abscessus_ATCC_1997 --------MAFDNPFLDSAGHIKFPEDGSIVGHVEGFAQSQGESLAYRFL
Mflv_1168|M.gilvum_PYR-GCK DFSTERDGVERDLHWADFSTRNKAVGARLQQVTEPGDRVAILCPQNLDYL
Mvan_5641|M.vanbaalenii_PYR-1 DFSTERDGIARDLTWADFGARNRAVAARLQQVTEPGDRVAILCPQNLEYL
MSMEG_6393|M.smegmatis_MC2_155 DFSTERDGVPRDLTWAQFSARNRAVAARLQQVTQPGDRVAILCPQNLDYL
TH_1609|M.thermoresistible__bu --------------------------------------------------
MUL_4984|M.ulcerans_Agy99 DFSTERDGVERDILWSEFSARNRAVGARLQQVTQPGDRIAILCPQNLDYL
MMAR_5365|M.marinum_M DFSTERDGVERDILWSEFSARNRAVGARLQQVTQPGDRIAILCPQNLDYL
Mb3831c|M.bovis_AF2122/97 DFSTERDGVARDILWSDFSARNRAVGARLQQVTQPGDRVAILCPQNLDYL
Rv3801c|M.tuberculosis_H37Rv DFSTERDGVARDILWSDFSARNRAVGARLQQVTQPGDRVAILCPQNLDYL
MLBr_00100|M.leprae_Br4923 DFSTGRDGVARDILWSEFSARNRAVGARLQQVTQPGDRIAILCPQNLDYL
MAV_0217|M.avium_104 DFSTERDGVACDISWSEFSARNRAVGARLQQVTEPGDRIAVLCPQNLDYL
MAB_0179|M.abscessus_ATCC_1997 DFSTERDGAYIDITWAQFGARNRAVAARLQQVTKPGDRVAILAPQSLDYL
Mflv_1168|M.gilvum_PYR-GCK VAFFGTLYSGRIAVPLFDPNEPGHVGRLHAVLDDCTPSAILTTTAAAEGV
Mvan_5641|M.vanbaalenii_PYR-1 VAFFGTLYSGRIAVPLFDPNEPGHVGRLHAVLDNCTPSAILTTTEAAEGV
MSMEG_6393|M.smegmatis_MC2_155 VAFFGALYAGRIAVPLFDPSEPGHVGRLHAVLDNCHPSAILTTTEAAEGV
TH_1609|M.thermoresistible__bu -------------------------------------------------V
MUL_4984|M.ulcerans_Agy99 ISFFGALYSGRIAVPLFDPAEPGHVGRLHAVLDDCTPSTILTSTDSAEGV
MMAR_5365|M.marinum_M ISFFGALYSGRIAVPLFDPAEPGHVGRLHAVLDDCTPSTILTTTDSAEGV
Mb3831c|M.bovis_AF2122/97 ISFFGALYSGRIAVPLFDPAEPGHVGRLHAVLDDCAPSTILTTTDSAEGV
Rv3801c|M.tuberculosis_H37Rv ISFFGALYSGRIAVPLFDPAEPGHVGRLHAVLDDCAPSTILTTTDSAEGV
MLBr_00100|M.leprae_Br4923 ISFFGALYSGRIAVPLFDPAEPGHVGRLHAVLDDCTPSTILTTTDSAEGV
MAV_0217|M.avium_104 IALFGALYAGRIAVPLFDPSEPGHVGRLHAVLDDCTPSTILTTTEAAEGV
MAB_0179|M.abscessus_ATCC_1997 VAHFGALYAGAISVPLFDPSEAGHAGRLHAVLDDCQPSAVLTTTDCAEGV
*
Mflv_1168|M.gilvum_PYR-GCK RKFFRTRPAKDRPRVIAVDAVPDEVGATWEP-VDVQHDTIAYLQYTSGST
Mvan_5641|M.vanbaalenii_PYR-1 RKFFRTRPANQRPRVIAVDAVPVEVGATWEP-VDVTHDTIAYLQYTSGST
MSMEG_6393|M.smegmatis_MC2_155 RKFFRTRPANQRPRVIAVDAVPDDVASTWVNPDEPDETTIAYLQYTSGST
TH_1609|M.thermoresistible__bu RKFFRTRPAKERPRVIAVDAVPDEVGSTWELYTDVDENTIAYLQYTSGST
MUL_4984|M.ulcerans_Agy99 RKFIRSRSAKKRPRVIAVDAVPTEVASTWQQ-PEANELTTAYLQYTSGST
MMAR_5365|M.marinum_M RKFIRSRSAKERPRVIAVDAVPTEVASTWQQ-PEANELTTAYLQYTSGST
Mb3831c|M.bovis_AF2122/97 RKFIRARSAKERPRVIAVDAVPTEVAATWQQ-PEANEETVAYLQYTSGST
Rv3801c|M.tuberculosis_H37Rv RKFIRARSAKERPRVIAVDAVPTEVAATWQQ-PEANEETVAYLQYTSGST
MLBr_00100|M.leprae_Br4923 RKFIRGRFAKERTRVIAVDAVPNEVAATWQQ-PEANEETVAYLQYTSGST
MAV_0217|M.avium_104 RKFIRARSAKERPRVIAVDAVPNEVNSTWVP-PEADENTIAYLQYTSGST
MAB_0179|M.abscessus_ATCC_1997 RKFFRNRPPKERPRVIAVDAIPNDVGSTWVE-PVATKDTIAYLQYTSGST
***:* * .:.*.*******:* :* :** . * **********
Mflv_1168|M.gilvum_PYR-GCK RIPTGVQITHLNLATNVVQVIEALDAEEGD------RGVSWLPFFHDMGL
Mvan_5641|M.vanbaalenii_PYR-1 RIPTGVQITHLNLATNVVQVIEALQGEEGD------RGVSWLPFFHDMGL
MSMEG_6393|M.smegmatis_MC2_155 RIPTGVQITHLNLATNVVQVIEALEGEEGD------RGLSWLPFFHDMGL
TH_1609|M.thermoresistible__bu RIPTGVQITHLNLATNVVQVIEALEGEEGD------RGVTWLPFFHDMGL
MUL_4984|M.ulcerans_Agy99 RVPSGVQITHLNLPTNVLQVLNALEGQEGD------RGVSWLPFFHDMGL
MMAR_5365|M.marinum_M RVPSGVQITHLNLPTNVLQVLNALEGQEGD------RGVSWLPFFHDMGL
Mb3831c|M.bovis_AF2122/97 RIPSGVQITHLNLPTNVVQVLNALEGQEGD------RGVSWLPFFHDMGL
Rv3801c|M.tuberculosis_H37Rv RIPSGVQITHLNLPTNVVQVLNALEGQEGD------RGVSWLPFFHDMGL
MLBr_00100|M.leprae_Br4923 RIPSGVQITHLNLPTNVLQVLNSLQGQEGD------RGVSWLPFFHDMGL
MAV_0217|M.avium_104 RTPTGVEITHLNLPTNVLQVLNGLEGKEGD------RGLSWLPFFHDMGL
MAB_0179|M.abscessus_ATCC_1997 RAPAGVQITHLNVATNLLQLIDALSAQEGAQGHEALRGCTWLPFFHDMGL
* *:**:*****:.**::*:::.*..:** ** :**********
Mflv_1168|M.gilvum_PYR-GCK ITVLLSPMLGHHISFMTPAAFVRRPGRWIRAMARKPDDTG--GVISVAPN
Mvan_5641|M.vanbaalenii_PYR-1 ITVLLPTVIGHYVEFMTPAAFVRRPGRWIRAMARKEGDTG--GTISVAPN
MSMEG_6393|M.smegmatis_MC2_155 ITALLAPMIGHYFTFMTPAAFVRRPERWIRELARKEGDTG--GTISVAPN
TH_1609|M.thermoresistible__bu ITVMLSPMIGHFVTFMTPAAFVRRPGRWIRELARKEGDTG--MTYSVAPN
MUL_4984|M.ulcerans_Agy99 ITVLLASVLGHSFTFMTPAAFVRRPGRWIRELARKPGETG--GTFSAAPN
MMAR_5365|M.marinum_M ITVLLASVLGHSFTFMTPAAFVRRPGRWIRELARKPGETG--GTFSAAPN
Mb3831c|M.bovis_AF2122/97 ITVLLASVLGHSFTFMTPAAFVRRPGRWIRELARKPGETG--GTFSAAPN
Rv3801c|M.tuberculosis_H37Rv ITVLLASVLGHSFTFMTPAAFVRRPGRWIRELARKPGETG--GTFSAAPN
MLBr_00100|M.leprae_Br4923 ITVLLPSVLGHSFTFMTPAAFVRRPGRWIRELARKPEDTG--GTFSAAPN
MAV_0217|M.avium_104 ITAMLSPVLGHNFTFMTPAAFVRRPGRWIREMARKPDDAPDCEVFTVAPN
MAB_0179|M.abscessus_ATCC_1997 ITVMVPSMLGKHMTVMSPAAFIRRPLRWLREMAVKGEDRG--GSFSALPN
**.::..::*: . .*:****:*** **:* :* * : :. **
Mflv_1168|M.gilvum_PYR-GCK FAFDHAAARALPKDDEPPLDLSNIKCILNGSEPISAATVRRFNEAFSPYG
Mvan_5641|M.vanbaalenii_PYR-1 FAFDHAAARGVPKDGEEPLDLSNVKAVLNGSEPISAATVRRFNEAFGPFG
MSMEG_6393|M.smegmatis_MC2_155 FAFDHAAARGVPKPGSPPLDLSNVKAVLNGSEPISAATVRRFNEAFGPFG
TH_1609|M.thermoresistible__bu FAFDHAAARGVPKDGEPPLDLSNVKAILNGSEPISAATVRRFNEAFGPFG
MUL_4984|M.ulcerans_Agy99 FAFEHAAMRGVPRDDEPPLDLSNVKGILNGSEPVSPASMRKFFKAFEPYG
MMAR_5365|M.marinum_M FAFEHAAMRGVPRDDEPPLDLSNVKGILNGSEPVSPASMRKFFKAFEPYG
Mb3831c|M.bovis_AF2122/97 FAFEHAAVRGVPRDDEPPLDLSNVKGILNGSEPVSPASMRKFFEAFAPYG
Rv3801c|M.tuberculosis_H37Rv FAFEHAAVRGVPRDDEPPLDLSNVKGILNGSEPVSPASMRKFFEAFAPYG
MLBr_00100|M.leprae_Br4923 FAFEHAAMRGLPRDDELPLDLSNVKGILNGSEPVSPASMRKFFEAFAPYG
MAV_0217|M.avium_104 FAFEHAAVRGVPKEGEPPLDLSNVKGILNGSEPVSPSSMRKFYEAFKPYG
MAB_0179|M.abscessus_ATCC_1997 FAFEHCALRGVPKEGEPPLDLSNIHSIINGSEPVSTASLKKFCDAFEPYG
***:*.* *.:*: .. ******:: ::*****:*.:::::* .** *:*
Mflv_1168|M.gilvum_PYR-GCK FKPEAIKPSYGLAEATLFVSTPPIHEHPK-IISVDRDELNSHRFVQVPDD
Mvan_5641|M.vanbaalenii_PYR-1 FQPQAIKPSYGLAEATLFVSTTPSNESPK-IISVDRDELNAHRFVEVPDD
MSMEG_6393|M.smegmatis_MC2_155 FPPKAIKPSYGLAEATLFVSTTPSAEEPK-IITVDRDQLNSGRIVEVDAD
TH_1609|M.thermoresistible__bu FQPKAIKPSYGLAEATLFVSTTPMGEPPK-IVTVDREELNNHRFVPVPED
MUL_4984|M.ulcerans_Agy99 LRETAVKPSYGLAEATLFVSTTPMDEVPT-VIHVDRDELNKQRFVEVAAD
MMAR_5365|M.marinum_M LRETAVKPSYGLAEATLFVSTTPMDEVPT-VIHVDRDELNKQRFVEVAAD
Mb3831c|M.bovis_AF2122/97 LKQTAVKPSYGLAEATLFVSTTPMDEVPT-VIHVDRDELNNQRFVEVAAD
Rv3801c|M.tuberculosis_H37Rv LKQTAVKPSYGLAEATLFVSTTPMDEVPT-VIHVDRDELNNQRFVEVAAD
MLBr_00100|M.leprae_Br4923 LKQTAVKPSYGLAEATLFVSTTPMDESPT-VIHVDRYELNNQRFVEVPAD
MAV_0217|M.avium_104 LRETAIKPSYGLAEATLFVSTTPMDQAPT-VIHVDRAELNKQRFVEVPAD
MAB_0179|M.abscessus_ATCC_1997 FDPKAIHPSYGMAEATLFVSSTIWNTEPARVLHVDRTELNAGRIVQVAPG
: *::****:********:. * :: *** :** *:* * .
Mflv_1168|M.gilvum_PYR-GCK SPKAVAQAGAGQIGIAEWAVIVDNDTATELPDGQIGEIWISGQNMGTGYW
Mvan_5641|M.vanbaalenii_PYR-1 SPKAVAQAGAGKVGVAEWAVIVDAETASELPDGQIGEIWISGQNMGTGYW
MSMEG_6393|M.smegmatis_MC2_155 SPKAVAQASAGKVGIAEWAVIVDAESATELPDGQVGEIWISGQNMGTGYW
TH_1609|M.thermoresistible__bu SPNAVPQASAGKVGVAEWAVIVDHETASELPDGQIGEIWISGQNMGTGYW
MUL_4984|M.ulcerans_Agy99 APNAVAQVSAGKVGVDEWAVIVDTETASELPDGQIGEIWLHGNNLGIGYW
MMAR_5365|M.marinum_M APNAVAQVSAGKVGVDEWAVIVDTETASELPDGQIGEIWLHGNNLGIGYW
Mb3831c|M.bovis_AF2122/97 APNAVAQVSAGKVGVSEWAVIVDADTASELPDGQIGEIWLHGNNLGTGYW
Rv3801c|M.tuberculosis_H37Rv APNAVAQVSAGKVGVSEWAVIVDADTASELPDGQIGEIWLHGNNLGTGYW
MLBr_00100|M.leprae_Br4923 APNAVAQVSAGKVGVSEWAIIVDADTASELPDGQIGEIWLHGNNLGTGYW
MAV_0217|M.avium_104 APNAVAQVSAGVIGVDEWAVIVDPETASELPDGHIGEIWLHGNNMGIGYW
MAB_0179|M.abscessus_ATCC_1997 AENSLTQVSSGRMARDEWAVIVDGESASEVPDGQIGQIWLHGNNIGSGYW
: :::.*..:* :. ***:*** ::*:*:***::*:**: *:*:* ***
Mflv_1168|M.gilvum_PYR-GCK NKPEETIATFQNLLKSRTTPSHAEGADADATWVRTGDLGAYHDGELFITG
Mvan_5641|M.vanbaalenii_PYR-1 NKPEETIATFHNTLKSRTTPSHAQGATDDATWVRTGDYGAYHDGELYITG
MSMEG_6393|M.smegmatis_MC2_155 GKPEESVATFQNILKSRTNPSHAEGATDDATWVRTGDYGAFYDGDLYITG
TH_1609|M.thermoresistible__bu GKPEETVATFQNILKSRTDPSHAEGAPDDATWVRTGDYGAYYDGDLYITG
MUL_4984|M.ulcerans_Agy99 GKEEESAQTFRNILKSRVPESHAEGAPDDGLWVRTGDYGTYFKGHLYIAG
MMAR_5365|M.marinum_M GKEEESAQTFRNILKSRVPESHAEGAPDDGLWVRTGDYGTYFKGHLYIAG
Mb3831c|M.bovis_AF2122/97 GKEEESAQTFKNILKSRISESRAEGAPDDALWVRTGDYGTYFKDHLYIAG
Rv3801c|M.tuberculosis_H37Rv GKEEESAQTFKNILKSRISESRAEGAPDDALWVRTGDYGTYFKDHLYIAG
MLBr_00100|M.leprae_Br4923 GKEEESAQTFNNILKSRISESRAEGASDDASWVRTGDYGTYFKDHLYIAG
MAV_0217|M.avium_104 GKEEETNEVFRNILKSRISQSHAEGAPDDAMWVKTGDYGTYYKGHLYIAG
MAB_0179|M.abscessus_ATCC_1997 GRPEETREAFQNTLKSRIPASHAEGAPDDANWLNTGDLGVWVDGELYITG
.: **: .*.* **** *:*:** *. *:.*** *.: ...*:*:*
Mflv_1168|M.gilvum_PYR-GCK RVKDLVIIDGRNHYPQDLEYSAQEATKALRTGFVAAFSVPANQLPDEVFD
Mvan_5641|M.vanbaalenii_PYR-1 RVKDLVIIDGRNHYPQDLEYSAQEATKALRTGYVAAFSVPANQLPDEVFD
MSMEG_6393|M.smegmatis_MC2_155 RVKDLVIIDGRNHYPQDLEYSAQEASKAIRTGYVAAFSVPANQLPDEVFE
TH_1609|M.thermoresistible__bu RVKDLVIIDGRNHYPQDLEHSAQEATKALRTGFVAAFSVPANQLPKEVFE
MUL_4984|M.ulcerans_Agy99 RIKDLVIIDGRNHYPQDLEYTAQESTKALRVGYVAAFSVPANQLPQKVFD
MMAR_5365|M.marinum_M RIKDLVIIDGRNHYPQDLEYTAQESTKALRVGYVAAFSVPANQLPQKVFD
Mb3831c|M.bovis_AF2122/97 RIKDLVIIDGRNHYPQDLECTAQESTKALRVGYVAAFSVPANQLPQTVFD
Rv3801c|M.tuberculosis_H37Rv RIKDLVIIDGRNHYPQDLECTAQESTKALRVGYAAAFSVPANQLPQTVFD
MLBr_00100|M.leprae_Br4923 RIKDLVIIDGRNHYPQDLEYTAQESTKALRVGYVAAFSVPANQLAQVVLE
MAV_0217|M.avium_104 RIKDLVIIDGRNHYPQDLEYSAQEASKALRTGYVAAFSVPANQLPKEVFD
MAB_0179|M.abscessus_ATCC_1997 RVKDLIIVDGRNHYPQDIEYTAQEANKALRPGYVAAFSVPANQLPQVVFD
*:***:*:*********:* :***:.**:* *:.**********.. *::
Mflv_1168|M.gilvum_PYR-GCK NAHAGIKRDPDDSSEQLVIVGERAPGAHKLDMGPIIDDIRAAIAVRHGVT
Mvan_5641|M.vanbaalenii_PYR-1 DSHAGLKRDPDDTSEQLVIVGERAPGAHKLDMGPIADDIRAAIAVRHGVT
MSMEG_6393|M.smegmatis_MC2_155 NAHSGIKRDPDDTSEQLVIVAERAPGAHKLDIGPITDDIRAAIAVRHGVT
TH_1609|M.thermoresistible__bu NAHSGIKYDPEDTSEQLVIVGERAPGAHKLDIGPITDDIRAAIAVRHGVT
MUL_4984|M.ulcerans_Agy99 DPHAGLSFDPEDTSEQLVIVGERAAGTHKLEYQPIADDIRAAIAVGHGVT
MMAR_5365|M.marinum_M DPHAGLSFDPEDTSEQLVIVGERAAGTHKLEYQPIADDIRAAIAVGHGVT
Mb3831c|M.bovis_AF2122/97 DSHAGLKFDPEDTSEQLVIVGERAAGTHKLDHQPIVDDIRAAIAVGHGVT
Rv3801c|M.tuberculosis_H37Rv DSHAGLKFDPEDTSEQLVIVGERAAGTHKLDHQPIVDDIRAAIAVGHGVT
MLBr_00100|M.leprae_Br4923 DPHVGLKFDAEDTSEQLVIVGERAAGTHKLDYQPIVDDIRAAIAVRHGVA
MAV_0217|M.avium_104 NPHTGLKYDPDDTSEQLVIVAERAPGTHKLDYQPIADDIRAAIAVRHGVT
MAB_0179|M.abscessus_ATCC_1997 NPHAGLKYDPEDSSEQLVIIGERSVGGHKSDNQAVGDDVRAAVAVRHGVT
:.* *:. *.:*:******:.**: * ** : .: **:***:** ***:
Mflv_1168|M.gilvum_PYR-GCK VRDVLLTPAGAIPRTSSGKIGRRACRSAYLDGSLRSGK-VANAFPDEVD
Mvan_5641|M.vanbaalenii_PYR-1 VRDVLLTPAGAIPRTSSGKIGRRACRSAYLDGSLRAGK-VANAFPDETD
MSMEG_6393|M.smegmatis_MC2_155 VRDVLLTAAGAIPRTSSGKIGRRACRAAYLDGSLRAGK-VANDFPDATD
TH_1609|M.thermoresistible__bu VRDVLLTPAGAIPRTSSGKIGRRACRAAYLDGSLRAGK-VANAFPDATD
MUL_4984|M.ulcerans_Agy99 VRDVLLVSAGTIPRTSSGKIGRRACRTAYIDGSLRSGVSSPTVFATGS-
MMAR_5365|M.marinum_M VRDVLLVSAGTIPRTSSGKIGRRACRTAYIDGSLRSGVSSPTVFATGS-
Mb3831c|M.bovis_AF2122/97 VRDVLLVSAGTIPRTSSGKIGRRACRAAYLDGSLRSGVGSPTVFATSD-
Rv3801c|M.tuberculosis_H37Rv VRDVLLVSAGTIPRTSSGKIGRRACRAAYLDGSLRSGVGSPTVFATSD-
MLBr_00100|M.leprae_Br4923 VRDVLLVPAGTIPRTSSGKIGRRACRAAYIDGSLRSGASSPTVFAAEF-
MAV_0217|M.avium_104 VRDLLLVQAGTIPRTSSGKIGHRACRAAYLDGSLRSGVGSPTAFANSTD
MAB_0179|M.abscessus_ATCC_1997 VRDVLLVPAGSIARTSSGKIARSAARTSYLDGSLRGGY-QQTAFPDDRQ
***:**. **:*.*******.: *.*::*:*****.* . *.