For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDSLRSTPMTAPPQETSVVAGERAVPVVSPGPLVPIADFGPVDRLRGWVVTGVITLLAAVTRFLNLGSP TDAGTPIFDEKHYAPQAWQVLNNHGIEDNPGFGLVVHPPVGKQMIAIGEALFGYTGLGWRFSGALLGVVM VALVVRIVRRITRSTLVGAIAGLLLICDGVSFVAARTALLDGFLTFFVVAAFGALIVDRDQVRARMHVAM LQGRIDETAWGPRLGVRWWRFGAGVLLGLAFATKWSGLYFIAFFGAMSLVFDVAARRQYRVPRPWLGMLR RDLIPTGYALALIPFGVYLASYAGWFASETAIDRHQVGLTIGPDSVVPLPDAIRSLWYSTAKAFHFHATL TNSAGNHHPWESKPWSWPMSLRPVLYAIDQQNVPGCGPQSCVKAEMLVGTPAMWWLAVPVLLYAAWRMFV RRDWRYAVVLVGYCAGWLPWFADIDRQMYFFYAATMAPFLVMAIALILGDILHQPGQGKERHTLGLIFVC CYIALVATNFAWLFPVLTGLPISQQTWDMQMWLPSWR
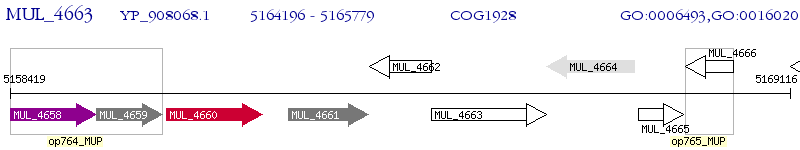
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4663 | - | - | 100% (527) | hypothetical protein MUL_4663 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1029c | - | 0.0 | 84.86% (502) | hypothetical protein Mb1029c |
| M. gilvum PYR-GCK | Mflv_1924 | - | 0.0 | 77.80% (518) | glycosyl transferase family protein |
| M. tuberculosis H37Rv | Rv1002c | - | 0.0 | 85.06% (502) | hypothetical protein Rv1002c |
| M. leprae Br4923 | MLBr_00192 | - | 0.0 | 77.84% (510) | hypothetical protein MLBr_00192 |
| M. abscessus ATCC 19977 | MAB_1122c | - | 0.0 | 66.79% (521) | hypothetical protein MAB_1122c |
| M. marinum M | MMAR_4491 | - | 0.0 | 99.23% (518) | hypothetical protein MMAR_4491 |
| M. avium 104 | MAV_1126 | - | 0.0 | 83.76% (511) | dolichyl-phosphate-mannose-protein mannosyltransferase |
| M. smegmatis MC2 155 | MSMEG_5447 | - | 0.0 | 73.83% (512) | dolichyl-phosphate-mannose-protein mannosyltransferase |
| M. thermoresistible (build 8) | TH_1147 | - | 0.0 | 74.71% (518) | CONSERVED MEMBRANE PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4809 | - | 0.0 | 76.66% (527) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1924|M.gilvum_PYR-GCK -----------MTAPTEELSRSGRAVPVISPGPVVPVADFHPLDRLQGWA
Mvan_4809|M.vanbaalenii_PYR-1 -----------MTAPTDQLERSGRTVPVISPGPVAPVADFGPVDRLQGWA
MSMEG_5447|M.smegmatis_MC2_155 -----------MTALDTDTPTAGRSAPLISPGPVIPPPDFGPLDRAQGWA
TH_1147|M.thermoresistible__bu -----------VTAPAADPVVTQRPVPVISPGPLVPVADFGPIDRLQGWA
MUL_4663|M.ulcerans_Agy99 MTDSLRSTPMTAPPQETSVVAGERAVPVVSPGPLVPIADFGPVDRLRGWV
MMAR_4491|M.marinum_M ---------MTAPPQETSVVAGERAVPVVSPGPLVPIADFGPVDRLRGWV
Mb1029c|M.bovis_AF2122/97 ------------------------MVPVVSPGPLVPVADFGPLDRLRGWI
Rv1002c|M.tuberculosis_H37Rv ------------------------MVPVVSPGPLVPVADFGPLDRLRGWI
MAV_1126|M.avium_104 --------------MHTDGAETERVVPVVSPGPLVPAADFGPTDDVRGWV
MLBr_00192|M.leprae_Br4923 ---------------------------MISPGPLVPVEDFGPTDDVRGWI
MAB_1122c|M.abscessus_ATCC_199 -----------MTAPATLPPTSAAESDLQSPGPRIPPADFGPTDRFRGWV
: **** * ** * * :**
Mflv_1924|M.gilvum_PYR-GCK MTAVITALAAVTRFLNLGSPTDAGTPIFDEKHYAPQAWQMVHNKGLEDNP
Mvan_4809|M.vanbaalenii_PYR-1 MTAVITALAAVTRFLNLGSPTDAGTPIFDEKHYAPQAWQMVHNGGVEDNP
MSMEG_5447|M.smegmatis_MC2_155 MTAIITALAAITRFLNLGSPTDAGTPIFDEKHYAPQAWQVLHNDGVEDNP
TH_1147|M.thermoresistible__bu MTAVIAALATLTRFLNLGSPTDAGTPIFDEKHYAPQAWQILHNNGVEDNP
MUL_4663|M.ulcerans_Agy99 VTGVITLLAAVTRFLNLGSPTDAGTPIFDEKHYAPQAWQVLNNHGIEDNP
MMAR_4491|M.marinum_M VTGVITLLAAVTRFLNLGSPTDAGTPIFDEKHYAPQAWQVLNNHGIEDNP
Mb1029c|M.bovis_AF2122/97 VTGLITLLATVTRFLNLGSLTDAGTPIFDEKHYAPQAWQVLNNHGVEDNP
Rv1002c|M.tuberculosis_H37Rv VTGLITLLATVTRFLNLGSLTDAGTPIFDEKHYAPQAWQVLNNHGVEDNP
MAV_1126|M.avium_104 VTGVVTLVAAVTRFVNLGSPTDGGTPVFDEKHYAPQAWQVLHNHGVEDNP
MLBr_00192|M.leprae_Br4923 ATGVITVLATGTRFINLSSPTDAGTPIFDEKHYAPQAWQLLHNHGVEDNP
MAB_1122c|M.abscessus_ATCC_199 MTAVITAIAALTRFINLGALTDRGTPVFDEKHYVPQGWQVVTNNGVEDNP
*.::: :*: ***:**.: ** ***:******.**.**:: * *:****
Mflv_1924|M.gilvum_PYR-GCK GYGLVVHPPVGKQLIAIGEAIFGYNGLGWRFSGAVCGVILVLLVVRIARR
Mvan_4809|M.vanbaalenii_PYR-1 GYGLVVHPPVGKQLIAIGEAIFGYNGLGWRFSGAVCGVVLVLLVVRIARR
MSMEG_5447|M.smegmatis_MC2_155 GYGLVVHPPVGKQLIAIGEWLFGYNGLGWRFSGAVCGVIIVMLVTRIARR
TH_1147|M.thermoresistible__bu GYGLVVHPPVGKQLIAIGEALFGYNALGWRFSGAVCGVLTIVLVARIARR
MUL_4663|M.ulcerans_Agy99 GFGLVVHPPVGKQMIAIGEALFGYTGLGWRFSGALLGVVMVALVVRIVRR
MMAR_4491|M.marinum_M GFGLVVHPPVGKQMIAIGEALFGYTGLGWRFSGALLGVVMVALVVRIVRR
Mb1029c|M.bovis_AF2122/97 GYGLVVHPPVGKQLIAIGEAIFGYNGFGWRFTGALLGVLLVALVVRIVRR
Rv1002c|M.tuberculosis_H37Rv GYGLVVHPPVGKQLIAIGEAIFGYNGFGWRFTGALLGVVLVALVVRIVRR
MAV_1126|M.avium_104 GFGLVVHPPVGKQLIAIGEALFGYNGVGWRFTGALLGVVMVALVVRIVRR
MLBr_00192|M.leprae_Br4923 GFGLVVHPPIGKQLIAIGEAIFGYNGVGWRFTGALLGVIMVALVMRIVRR
MAB_1122c|M.abscessus_ATCC_199 GYGLVVHPPIGKQMIAIGEWLFGYDPFGWRFAAALSGTILVMLVARIARR
*:*******:***:***** :*** .****:.*: *.: : ** **.**
Mflv_1924|M.gilvum_PYR-GCK ISRSTLVGGIAGLLLIADGVSFVTARTALLDGFLTLFVVAAFGALIVDRD
Mvan_4809|M.vanbaalenii_PYR-1 ISRSTLVGGIAGLLLIADGVSFVTARTALLDGFLTVFVVAAFGALIVDRD
MSMEG_5447|M.smegmatis_MC2_155 ISRSTLVGAIAGLLIIADGVSFVSSRTALLDVFLVMFAVAAFACLMVDRD
TH_1147|M.thermoresistible__bu ISRSTLVGGIAGLLLIADGVSFVSSRTALLDVFLVTFVVAAFGCLMVDRD
MUL_4663|M.ulcerans_Agy99 ITRSTLVGAIAGLLLICDGVSFVAARTALLDGFLTFFVVAAFGALIVDRD
MMAR_4491|M.marinum_M ITRSTLVGAIAGLLLICDGVSFVAARTALLDGFLTFFVVAAFGALIVDRD
Mb1029c|M.bovis_AF2122/97 ISRSTLVGAIAGVLLICDGVSFVTARTALLDGFLTFFVVAAFGALIVDRD
Rv1002c|M.tuberculosis_H37Rv ISRSTLVGAIAGVLLICDGVSFVTARTALLDGFLTFFVVAAFGALIVDRD
MAV_1126|M.avium_104 ISRSTLVGAIAGVLVICDGVSFVASRTALLDGILTFFVVAAFGALIVDRD
MLBr_00192|M.leprae_Br4923 ITRSTLIGTIAGLLVICDGVSFVTARTALLDGFLTFFVVATFGALIVDRD
MAB_1122c|M.abscessus_ATCC_199 ISRSTMVGAIAGVLLIADGVSFVTARTALLDVYQALFVVAAFGALIVDRD
*:***::* ***:*:*.******::****** . *.**:*..*:****
Mflv_1924|M.gilvum_PYR-GCK QIRERMHVAFLEGRIDETPWGPRLGVRWWRFGAGVLLGLACATKWSGLYF
Mvan_4809|M.vanbaalenii_PYR-1 QIRERMHIAFVEGRIAETPWGPRLGVRWWRFGAGVLLGLACATKWSGLYF
MSMEG_5447|M.smegmatis_MC2_155 QVRERMYHAFLDGRIAETRWGTRLGVRWWRFGAGVLLGLACATKWSGLYF
TH_1147|M.thermoresistible__bu QVRARMHNALVEGRIAETPWGPRLGVRWWRFGAGVLLGLACATKWSGLYF
MUL_4663|M.ulcerans_Agy99 QVRARMHVAMLQGRIDETAWGPRLGVRWWRFGAGVLLGLAFATKWSGLYF
MMAR_4491|M.marinum_M QVRARMHVAMLQGRIGETAWGPRLGVRWWRFGAGVLLGLAFATKWSGLYF
Mb1029c|M.bovis_AF2122/97 QVRERMHIALLAGRSAATVWGPRVGVRWWRFGAGVLLGLACATKWSGVYF
Rv1002c|M.tuberculosis_H37Rv QVRERMHIALLAGRSAATVWGPRVGVRWWRFGAGVLLGLACATKWSGVYF
MAV_1126|M.avium_104 QVRQRLHTALLQGRSADTVWGPRLGVRWWRFGAGVLLGLACGTKWSGLYF
MLBr_00192|M.leprae_Br4923 QVRQRMHTAITEGRSTETVWGPRLGVRWWRFLGGILLGLACATKWSGLYF
MAB_1122c|M.abscessus_ATCC_199 QVRQRLYNAYLEGRIGDTPWGPRLGVRWWRFGAGILLGLACGTKWSGLYF
*:* *:: * ** * **.*:******* .*:***** .*****:**
Mflv_1924|M.gilvum_PYR-GCK VMFFGLMTVAYTVAARKQYRVPRPWLGTLGRDVGPSLYALVLIPFGVYLA
Mvan_4809|M.vanbaalenii_PYR-1 VMFFGVMTVAFDIAARRQYRVPRPWLGTFRRDVGPSLYALVLIPFGVYLA
MSMEG_5447|M.smegmatis_MC2_155 VLFFGVMTLVFDAIARKQYHVPHPWRGMLRRDLGPAAYVFGLIPFAVYLA
TH_1147|M.thermoresistible__bu VVFFGLMTLAFDVVARRQYHVPRPWLGVLRRDVGPAAYALGLIPAGVYLA
MUL_4663|M.ulcerans_Agy99 IAFFGAMSLVFDVAARRQYRVPRPWLGMLRRDLIPTGYALALIPFGVYLA
MMAR_4491|M.marinum_M IAFFGAMSLAFDVAARRQYRVPRPWLGMLRRDLIPTGYALALIPFGVYLA
Mb1029c|M.bovis_AF2122/97 VLFFGAMALAFDVAARRQYQVQRPWLGTVRRDVLPSGYALGLIPFAVYLA
Rv1002c|M.tuberculosis_H37Rv VLFFGAMALAFDVAARRQYQVQRPWLGTVRRDVLPSGYALGLIPFAVYLA
MAV_1126|M.avium_104 VLFFAAMSLAFDVAARRQYQVTRPWLGVVRRDLLPTGYALGFIPAAVYLA
MLBr_00192|M.leprae_Br4923 VLFFGVMSLAFDVSSRRQYQVLNPWRGVLRRDVIPTVYALGLISFLVYIT
MAB_1122c|M.abscessus_ATCC_199 IAFFGVLCIAFDIAARRAYGVQRPWAGAAVRDLGPALYALLIIPFLVYMA
: **. : :.: :*: * * .** * **: *: *.: :*. **::
Mflv_1924|M.gilvum_PYR-GCK SYTPWFASETGVDRHEVGRSIGPDSVLPLPDAVRSLWHYTYAAYRFHSGL
Mvan_4809|M.vanbaalenii_PYR-1 SYAPWFASETAVDRYQVGRSIGPDSVLPIPDALRSLWHYTYAAYRFHSGL
MSMEG_5447|M.smegmatis_MC2_155 SYAPWFASETAVNRYEVGRSIGPDSILPIPDALRSLWHYTHAAYRFHSNL
TH_1147|M.thermoresistible__bu AFAPWFASETAIHRYAAGRVIGESSLLPIPDALRSLWHYQYGAFKFHSGL
MUL_4663|M.ulcerans_Agy99 SYAGWFASETAIDRHQVGLTIGPDSVVPLPDAIRSLWYSTAKAFHFHATL
MMAR_4491|M.marinum_M SYAGWFASETAIDRHQVGLTIGPDSVVPLPDAIRSLWYYTAKAFHFHATL
Mb1029c|M.bovis_AF2122/97 TYAPWFASETAIDRHAVGQAVGRNSVVPLPDAVRSLWHYTAKAFHFHAGL
Rv1002c|M.tuberculosis_H37Rv TYAPWFASETAIDRHAVGQAVGRNSVVPLPDAVRSLWHYTAKAFHFHAGL
MAV_1126|M.avium_104 GYAPWFASETAIDRHEVGQTIGPRSVLPLPDAIRSLWHYTAKAFQFHAGL
MLBr_00192|M.leprae_Br4923 SYAPWFASETAIDRHQVGQTIGRHGMIPLPDAVRSLWYYHAKTLQFHAGL
MAB_1122c|M.abscessus_ATCC_199 SYWSWFASETGINRHAEGRQIGVGGWMP--DALRSLWYYTHSVYTFHSTL
: ******.:.*: * :* . :* **:****: . **: *
Mflv_1924|M.gilvum_PYR-GCK TNADGNHHPWESKPWAWPMSLRPVLYAIDTENVSGCGAQSCVKAVMLVGT
Mvan_4809|M.vanbaalenii_PYR-1 TNADGNHHPWESKPWTWPMSLRPVLYAIDTENVSGCGAQSCVKAVMLVGT
MSMEG_5447|M.smegmatis_MC2_155 TNADGNHHPWESKPWTWPMSLRPVLYAIDNQDVPGCGAQSCVKAVMLVGT
TH_1147|M.thermoresistible__bu TNAAGNHHPWESKPWTWPMSLRPVLYAIDNQNVPGCGEASCVKAVMMVGT
MUL_4663|M.ulcerans_Agy99 TNSAGNHHPWESKPWSWPMSLRPVLYAIDQQNVPGCGPQSCVKAEMLVGT
MMAR_4491|M.marinum_M TNSAGNHHPWESKPWSWPMSLRPVLYAIDQQNVPGCGPQSCVKAEMLVGT
Mb1029c|M.bovis_AF2122/97 TNSAGNYHPWESKPWTWPMSLRPVLYAIDQQDVAGCGAQSCVKAEMLVGT
Rv1002c|M.tuberculosis_H37Rv TNSAGNYHPWESKPWTWPMSLRPVLYAIDQQDVAGCGAQSCVKAEMLVGT
MAV_1126|M.avium_104 TNSAGNYHPWESKPWSWPMSLRPVLYAIDEQNVPGCGAQSCVKAEMLVGT
MLBr_00192|M.leprae_Br4923 TNSAGNYHPWESKPWSWPMSLRPVLYAIDQQDVPGCGAQSCVKAEMLVGT
MAB_1122c|M.abscessus_ATCC_199 TNSNGNHHPWESKPWTWPMSLRPLLYAIEDKNISGCGTNSCVRAVLLVGT
**: **:********:*******:****: :::.*** ***:* ::***
Mflv_1924|M.gilvum_PYR-GCK PTMWFLAVPVLAWAVWRAFVKRDWRYAVLLVGYNAGFLPWFADIDRQMYF
Mvan_4809|M.vanbaalenii_PYR-1 PAMWFLAVPVLAWAVWRAFVKRDWRYAVLLVGYSAGFLPWFADIDRQMYF
MSMEG_5447|M.smegmatis_MC2_155 PAMWFIAVPVLGWALWRTVVRRDWRYGAVLVGYMAGFLPWFADIDRQMYF
TH_1147|M.thermoresistible__bu PAMWFIAVPVLGWALWRAFVKRDWRYAAVLVGYGAGYLPWFVDIDRQMYF
MUL_4663|M.ulcerans_Agy99 PAMWWLAVPVLLYAAWRMFVRRDWRYAVVLVGYCAGWLPWFADIDRQMYF
MMAR_4491|M.marinum_M PAMWWLAVPVLLYAAWRMFVRRDWRYAAVLVGYCAGWLPWFADIDRQMYF
Mb1029c|M.bovis_AF2122/97 PAMWWLAVPVLAYAGWRMFVRRDWRYAVVLVGYCAGWLPWFADIDRQMYF
Rv1002c|M.tuberculosis_H37Rv PAMWWLAVPVLAYAGWRMFVRRDWRYAVVLVGYCAGWLPWFADIDRQMYF
MAV_1126|M.avium_104 PAMWWLAVPVLVFALWRMLVRRDWRYAAVLVGYCAGWLPWFADIDRQMYF
MLBr_00192|M.leprae_Br4923 PAMWWLAVPVLVYATWRTVIQRDWRYAAVLVGYCAGWLPWFADIDRQMYL
MAB_1122c|M.abscessus_ATCC_199 PAIWWLAVPVLLWATWATVVRRDWRYAAALVGYCAGWLPWFANIDRQMYF
*::*::***** :* * .::*****.. **** **:****.:******:
Mflv_1924|M.gilvum_PYR-GCK FYASTMAPFLVLMIAMILGDILYKPGQN----------SERRTLGLLVVC
Mvan_4809|M.vanbaalenii_PYR-1 FYAATMAPFLVMIIAMILGDILYKPGQNTADVR-SSLSTERRTLGLLVVC
MSMEG_5447|M.smegmatis_MC2_155 FYATVMAPFLVLAIALILGDILYKPNQN----------PERRTLGLLTVC
TH_1147|M.thermoresistible__bu FYATTMAPFLVLAIALILGDILYKPTRN----------TERRTLALLVVS
MUL_4663|M.ulcerans_Agy99 FYAATMAPFLVMAIALILGDILHQPGQG----------KERHTLGLIFVC
MMAR_4491|M.marinum_M FYAATMAPFLVMAIALILGDILHQPGQG----------KERHTLGLIFVC
Mb1029c|M.bovis_AF2122/97 FYAATMAPFLVMGISLVLGDILYHPGQG----------SERRTLGLIVVC
Rv1002c|M.tuberculosis_H37Rv FYAATMAPFLVMGISLVLGDILYHPGQG----------SERRTLGLIVVC
MAV_1126|M.avium_104 FYAATMAPFLAMAIALICGDILYAPTATGMLRA----NSERRTLGLIAVC
MLBr_00192|M.leprae_Br4923 FYAATMAPFLVMGIALILGDILYPPWHDSSPRSPSTHSTERRTLRLIAVS
MAB_1122c|M.abscessus_ATCC_199 FYATPMVPFLVMMIAFIIGDFLRKPTDN----------PERKKLRMFIAT
***: *.***.: *::: **:* * **:.* :: .
Mflv_1924|M.gilvum_PYR-GCK IYVAVVITNFAWLYPVLTGLPISQHTWNMQIWLPSWR
Mvan_4809|M.vanbaalenii_PYR-1 FYVAMVITNFAWLYPILTGLPISQHTWNMQIWLPSWR
MSMEG_5447|M.smegmatis_MC2_155 FYVALVITNFAWMYPILTGLPISQTTWNLQIWLPSWR
TH_1147|M.thermoresistible__bu FYVALVLTNFAWLYPILTGLPISQSTWNMQIWLPSWR
MUL_4663|M.ulcerans_Agy99 CYIALVATNFAWLFPVLTGLPISQQTWDMQMWLPSWR
MMAR_4491|M.marinum_M CYIALVATNFAWLFPVLTGLPISQQTWDMQMWLPSWR
Mb1029c|M.bovis_AF2122/97 CYVALVVTNFAWLYPVLTGLPISQQTWNLEIWLPSWR
Rv1002c|M.tuberculosis_H37Rv CYVALVVTNFAWLYPVLTGLPISQQTWNLEIWLPSWR
MAV_1126|M.avium_104 CYVALVVTNFAWLFPVLTGLPISQQTWNMEIWLPSWR
MLBr_00192|M.leprae_Br4923 CYVALVITNFVWLYPVLTGLPISHQTWNMEIWLPSWR
MAB_1122c|M.abscessus_ATCC_199 FYLALVVTNFAWLYPILTGAAISPFMWNMEMWLPSWR
*:*:* ***.*::*:*** .** *::::******