For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADAKRPDPKRRSPASRPGKAGESARGRRTTSVSRPTQAANTGPAASRNTARTLREHVVEPIKRSFTESVE QRSDQRLGFTARRAAVLAAVVCVLTLTIAGPVRTYFAQRTEMEQLSATEAALRRQIADLEQRKGQLADPA YIAAQARERLGFVMPGDIPFQVQLPPGAVVAPQPGTDAAMPTKGDPWYTSLWHTIADAPHLPPAAAEPPA PEPEPEPGLPAPVPPEPTAPGG*
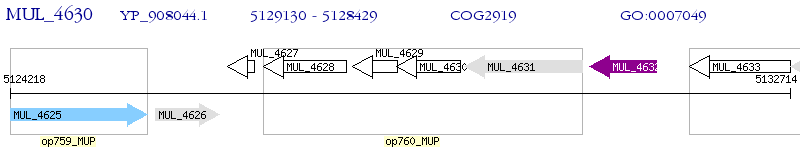
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4630 | - | - | 100% (233) | hypothetical protein MUL_4630 |
| M. ulcerans Agy99 | MUL_4514 | - | 2e-05 | 36.90% (84) | membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1052 | - | 6e-90 | 71.67% (233) | hypothetical protein Mb1052 |
| M. gilvum PYR-GCK | Mflv_1955 | - | 5e-66 | 55.87% (247) | septum formation initiator |
| M. tuberculosis H37Rv | Rv1024 | - | 6e-90 | 71.67% (233) | hypothetical protein Rv1024 |
| M. leprae Br4923 | MLBr_00256 | - | 6e-84 | 72.09% (215) | hypothetical protein MLBr_00256 |
| M. abscessus ATCC 19977 | MAB_1166 | - | 1e-52 | 53.02% (215) | hypothetical protein MAB_1166 |
| M. marinum M | MMAR_4461 | - | 1e-134 | 100.00% (233) | hypothetical protein MMAR_4461 |
| M. avium 104 | MAV_1165 | - | 3e-86 | 71.74% (230) | septum formation initiator subfamily protein, putative |
| M. smegmatis MC2 155 | MSMEG_5414 | - | 1e-68 | 59.31% (231) | septum formation initiator subfamily protein, putative |
| M. thermoresistible (build 8) | TH_0861 | - | 6e-48 | 65.75% (146) | POSSIBLE CONSERVED MEMBRANE PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4770 | - | 3e-65 | 56.35% (252) | septum formation initiator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1955|M.gilvum_PYR-GCK MPDAKRPDPKRRSSS----AKKAPASRPGK---SGDAGRAKPRAIAARRE
Mvan_4770|M.vanbaalenii_PYR-1 MPEANRPDPRRRSSAGDARSRKAPASRPGKPGKSGDAARGKPRG-AARRE
MSMEG_5414|M.smegmatis_MC2_155 MPDPKRPDPRR----------RAPAPRPGKPGKNGDANR--PRASQARRR
TH_0861|M.thermoresistible__bu --------------------------------------------------
MUL_4630|M.ulcerans_Agy99 MADAKRPDPKR----------RSPASRPGKAGESARGRRTTSVSR-----
MMAR_4461|M.marinum_M MADAKRPDPKR----------RSPASRPGKAGESARGRRTTSVSR-----
Mb1052|M.bovis_AF2122/97 MPEAKRPESKR----------RSPASRPGKAGDSVRGGRATKPS------
Rv1024|M.tuberculosis_H37Rv MPEAKRPESKR----------RSPASRPGKAGDSVRGGRATKPS------
MAV_1165|M.avium_104 MPAK--PDPKR----------RSPASRPGKAGDSVRRRRAAKPS------
MLBr_00256|M.leprae_Br4923 MSEAKRLDPKR----------RSPASRPGKAGDSVRGRRSTKPVAKLSVK
MAB_1166|M.abscessus_ATCC_1997 MADSKRRPASGRS--------RGPAPAARKRTPVKRPVQKVVRG------
Mflv_1955|M.gilvum_PYR-GCK SRSAEP-PPT----AREDAEFVERPDGDGDGAGSIRESILASAEAAAEQR
Mvan_4770|M.vanbaalenii_PYR-1 TRSADPRSPKELTGAAADPGFVDDT-GDEPGADSIRQSIIASAEQAAEQR
MSMEG_5414|M.smegmatis_MC2_155 AAEARP------------AQIEQAP--DESVAKAIAVQAQQQAELQSEQR
TH_0861|M.thermoresistible__bu --------------------------------------------------
MUL_4630|M.ulcerans_Agy99 PTQAAN--------TGPAASRNTARTLREHVVEPIKRSFTESVEQRSDQR
MMAR_4461|M.marinum_M PTQAAN--------TGPAASRNTARTLREHVVEPIKRSFTESVEQRSDQR
Mb1052|M.bovis_AF2122/97 -AKPST--------PAPHASRKTTRTPHEHIVEPIKRAITESVEKRSEQR
Rv1024|M.tuberculosis_H37Rv -AKPST--------PAPHASRKTTRTPHEHIVEPIKRAITESVEKRSEQR
MAV_1165|M.avium_104 -QTVT--------------ARKTARAQHEPAVEPIVRQAAESVEQRSEQR
MLBr_00256|M.leprae_Br4923 PSRTTP--------ASSHSGRNSTRMLTQHVVEPIRQSIIESRERRSDQQ
MAB_1166|M.abscessus_ATCC_1997 PARSGP-------------AAPAKATKSGGASSAVAASIERSAEHQSEQR
Mflv_1955|M.gilvum_PYR-GCK FGSAARRAAILAAVICVLTLTIAGPVRTYFSQRTEMKQLQASEAQLREQI
Mvan_4770|M.vanbaalenii_PYR-1 FGSAARRAAILAAVICVLTLTIAGPVRTYFAQRTEMKQLQASEAQLRDQI
MSMEG_5414|M.smegmatis_MC2_155 FGSAARRAAILAAVVCVLTLTIAGPVRTYFAQRTEMKQLKASEEQLRAQI
TH_0861|M.thermoresistible__bu ----------------VLTLTIAGPVRTYFAQRTEMMQLKAVQEQLREQI
MUL_4630|M.ulcerans_Agy99 LGFTARRAAVLAAVVCVLTLTIAGPVRTYFAQRTEMEQLSATEAALRRQI
MMAR_4461|M.marinum_M LGFTARRAAVLAAVVCVLTLTIAGPVRTYFAQRTEMEQLSATEAALRRQI
Mb1052|M.bovis_AF2122/97 LGFTARRAAILAAVVCVLTLTIARPVRTYFAQRAEMEQLAATEAMLRRQI
Rv1024|M.tuberculosis_H37Rv LGFTARRAAILAAVVCVLTLTIARPVRTYFAQRAEMEQLAATEAMLRRQI
MAV_1165|M.avium_104 LGFTARRAAVLAAVICVLTLTIAGPVRTYFAQRTEMNQLAAGEAALRRQI
MLBr_00256|M.leprae_Br4923 LGFTARRAAVLAAVVCVLTLTIAGPVRTYFAQHAEIEQLAATEATLRRQI
MAB_1166|M.abscessus_ATCC_1997 LGSTARRAAILAAVVCALTLTVAGPVRTYFSQQAEKSQLAAAEAQLRDQI
.****:* ******:*::* ** * : ** **
Mflv_1955|M.gilvum_PYR-GCK SELEEQKAKLADPVFIAAQARERLGFVMPGETPYQVQLPPGAAVAPNIPA
Mvan_4770|M.vanbaalenii_PYR-1 SELEQQKAKLADPVFIAAQARERLGFVMPGEIPYQVQLPPG-AAQPAAPP
MSMEG_5414|M.smegmatis_MC2_155 AELEQQKVKLADPVYIAAQARERLGFVMPGDTPFQVQLPPNAVVTQEPEP
TH_0861|M.thermoresistible__bu AELEEQKLKLADPAYIAAQARERLGFVMPGDIPYQVQLPPGAEVP-EP-P
MUL_4630|M.ulcerans_Agy99 ADLEQRKGQLADPAYIAAQARERLGFVMPGDIPFQVQLPPGAVVAPQPGT
MMAR_4461|M.marinum_M ADLEQRKGQLADPAYIAAQARERLGFVMPGDIPFQVQLPPGAVVAPQPGT
Mb1052|M.bovis_AF2122/97 ADLEEQQVKLADPAYIAAQARERLGFVMPGDIPFQVQLPSTPLAPPQPGS
Rv1024|M.tuberculosis_H37Rv ADLEEQQVKLADPAYIAAQARERLGFVMPGDIPFQVQLPSTPLAPPQPGS
MAV_1165|M.avium_104 ADLEQRKAKLADPAYIAAQARERLGFVKPGDIPFQVQLPPTAAEPTQPGS
MLBr_00256|M.leprae_Br4923 ADLEQQKGKLADSAYIAARARERLGFVMPGDVPFQVQLPSTAAVSSQPGG
MAB_1166|M.abscessus_ATCC_1997 SSLEDKKRRLADPAYIAAQARERLGFVMPGEVPFQVQLPNT-PVDVKPQG
:.**::: :***..:***:******** **: *:*****
Mflv_1955|M.gilvum_PYR-GCK EELLEPK-NNDPWYTSLWHTISDVPQGISPTA--PPVP---PGAP-APPP
Mvan_4770|M.vanbaalenii_PYR-1 DELAEAK-TNDPWYTSLWHTIADAPHGVSPMA--PPAP---PGVPGAPEP
MSMEG_5414|M.smegmatis_MC2_155 EKPGATVPAGQPWYTSLWHTIADQPHGVTPTI--PPAPP-VPPAPGEPAP
TH_0861|M.thermoresistible__bu DAHAAVAPSDEPWYTSLWRTIADEPHGPAPSP--GPAPD-DPSAP-VPDG
MUL_4630|M.ulcerans_Agy99 DAAMPTK--GDPWYTSLWHTIADAPH-LPPAAAEPPAPE-PEPEPGLPAP
MMAR_4461|M.marinum_M DAAMPTK--GDPWYTSLWHTIADAPH-LPPAAAEPPAPE-PEPEPGLPAP
Mb1052|M.bovis_AF2122/97 DAATATN--NEPWYTALWHTIADDPH-LPPAA--PPAP-----EPGRPGP
Rv1024|M.tuberculosis_H37Rv DAATATN--NEPWYTALWHTIADDPH-LPPAA--PPAP-----EPGRPGP
MAV_1165|M.avium_104 PSAKPVN--NDPWYTSLWHTIADAPH-LPPAN--PAVPQAPPAEPGPPGP
MLBr_00256|M.leprae_Br4923 RAAKPAN--NDPWYTSLWHNIADAPH-LPPGA-------------GTP-P
MAB_1166|M.abscessus_ATCC_1997 QGPVDTG---NPWYTNLWHNIADSPTVVPTPP--PAVP------PAPPAP
:**** **:.*:* * .. *
Mflv_1955|M.gilvum_PYR-GCK TAP--------------------------------GAP--------APGG
Mvan_4770|M.vanbaalenii_PYR-1 VAP--------------------------------GAP--------PSGG
MSMEG_5414|M.smegmatis_MC2_155 IPPAPVVDPADLEVVARQLGREPRGVLEIAYRCPNGEPGVVKTAPRLPDG
TH_0861|M.thermoresistible__bu PPPAPVEP------------------------VPPGEP--------MPGG
MUL_4630|M.ulcerans_Agy99 VP-------------------------------PEPTA---------PGG
MMAR_4461|M.marinum_M VP-------------------------------PEPTA---------PGG
Mb1052|M.bovis_AF2122/97 LPPAS----------------------------PNPEQ---------PGG
Rv1024|M.tuberculosis_H37Rv LPPAS----------------------------PNPEQ---------PGG
MAV_1165|M.avium_104 VPPPA----------------------------PNPAG---------PGG
MLBr_00256|M.leprae_Br4923 FS-LT----------------------------PLSTT---------SGG
MAB_1166|M.abscessus_ATCC_1997 HP------------------------------------------------
.
Mflv_1955|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4770|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5414|M.smegmatis_MC2_155 TPFPTLYYLTHPALTAAASRLESSGMMREMTERLAQDPELAAAYRRAHES
TH_0861|M.thermoresistible__bu --------------------------------------------------
MUL_4630|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4461|M.marinum_M --------------------------------------------------
Mb1052|M.bovis_AF2122/97 --------------------------------------------------
Rv1024|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1165|M.avium_104 --------------------------------------------------
MLBr_00256|M.leprae_Br4923 --------------------------------------------------
MAB_1166|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1955|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4770|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5414|M.smegmatis_MC2_155 YLAERDAIEPLGTTFTGGGMPDRVKCLHVVIAHSLAKGPGVNPFGDEALA
TH_0861|M.thermoresistible__bu --------------------------------------------------
MUL_4630|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4461|M.marinum_M --------------------------------------------------
Mb1052|M.bovis_AF2122/97 --------------------------------------------------
Rv1024|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1165|M.avium_104 --------------------------------------------------
MLBr_00256|M.leprae_Br4923 --------------------------------------------------
MAB_1166|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1955|M.gilvum_PYR-GCK ------------------
Mvan_4770|M.vanbaalenii_PYR-1 ------------------
MSMEG_5414|M.smegmatis_MC2_155 VLAAEPAMAGILERDTWV
TH_0861|M.thermoresistible__bu ------------------
MUL_4630|M.ulcerans_Agy99 ------------------
MMAR_4461|M.marinum_M ------------------
Mb1052|M.bovis_AF2122/97 ------------------
Rv1024|M.tuberculosis_H37Rv ------------------
MAV_1165|M.avium_104 ------------------
MLBr_00256|M.leprae_Br4923 ------------------
MAB_1166|M.abscessus_ATCC_1997 ------------------