For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRPAIKVGLSTASVYPLRAEAAFEYAARLGYDGVELMVWSESISQDIDAVRKLSRRYRVPVLSVHAPCLL ISQRVWGANPILKLDRSVRAAEELGAQTVVVHPPFRWQRRYAEGFSDEVAALEASSDVVVAVENMFPFRA DRFFGAGQSRERMRKRGGGPGAAISAFAPSYDPLDGDHAHYTLDLSHTSTAGTDALSMARRMGSGLMHLH LCDGSGLPADEHLVPGRGTEPTAEVCQMLAGRGFAGQVVLEVSTSSARSAAEREAMLSESLQFARTHLMR
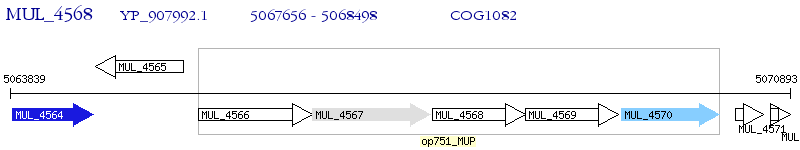
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4568 | - | - | 100% (280) | hypothetical protein MUL_4568 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0509 | - | 1e-149 | 91.79% (280) | hypothetical protein Mb0509 |
| M. gilvum PYR-GCK | Mflv_0077 | - | 1e-133 | 82.56% (281) | xylose isomerase domain-containing protein |
| M. tuberculosis H37Rv | Rv0498 | - | 1e-149 | 91.79% (280) | hypothetical protein Rv0498 |
| M. leprae Br4923 | MLBr_02432 | - | 1e-141 | 85.71% (280) | hypothetical protein MLBr_02432 |
| M. abscessus ATCC 19977 | MAB_4011c | - | 1e-126 | 77.94% (281) | hypothetical protein MAB_4011c |
| M. marinum M | MMAR_0824 | - | 1e-158 | 99.29% (280) | hypothetical protein MMAR_0824 |
| M. avium 104 | MAV_4654 | - | 1e-144 | 88.57% (280) | hypothetical protein MAV_4654 |
| M. smegmatis MC2 155 | MSMEG_0941 | - | 1e-131 | 81.85% (281) | AP endonuclease, family protein 2 |
| M. thermoresistible (build 8) | TH_0883 | - | 1e-135 | 82.92% (281) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_0835 | - | 1e-133 | 81.85% (281) | xylose isomerase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4568|M.ulcerans_Agy99 MRPAIKVGLSTASVYPLRAEAAFEYAARLGYDGVELMVWSESISQDIDAV
MMAR_0824|M.marinum_M MRPAIKVGLSTASVYPLRAEAAFEYAARLGYDGVELMVWSESISQDIDAV
Mb0509|M.bovis_AF2122/97 MRPAIKVGLSTASVYPLRAEAAFEYADRLGYDGVELMVWGESVSQDIDAV
Rv0498|M.tuberculosis_H37Rv MRPAIKVGLSTASVYPLRAEAAFEYADRLGYDGVELMVWGESVSQDIDAV
MLBr_02432|M.leprae_Br4923 MRPVIKVGLSTASVYPLRAEAAFEYAAKLGYDGVELMVWGESVSQDIDAV
MAV_4654|M.avium_104 MRPAIKVGLSTASVYPLRAEAAFEYAARLGYDGVELMVWAESASQDIAAV
Mflv_0077|M.gilvum_PYR-GCK MRPAIKVGLSTASVYPLKTEAAFEYAANLGYDGVELMVWAEAVSQDIDAI
TH_0883|M.thermoresistible__bu VRPAIKVGLSTASVYPLRTEAAFEYAARLGYDGVELMVWAESVSQDIDAV
Mvan_0835|M.vanbaalenii_PYR-1 MRPAIKVGLSTASVYPLRTEAAFEYAAKLGYDGVELMVWAEPVSQDVEAV
MSMEG_0941|M.smegmatis_MC2_155 MRPAIKVGLSTASVYPLRTEAAFEIAARLGYDGVELMVWAESVSQDIDAV
MAB_4011c|M.abscessus_ATCC_199 MRPAIKVGLSTASVYPLKTEAAFEYAARLGYDGVELMVWAEAVSQDVNAV
:**.*************::***** * .***********.*. ***: *:
MUL_4568|M.ulcerans_Agy99 RKLSRRYRVPVLSVHAPCLLISQRVWGANPILKLDRSVRAAEELGAQTVV
MMAR_0824|M.marinum_M RKLSRRYRVPVLSVHAPCLLISQRVWGANPILKLDRSVRAAEELGAQTVV
Mb0509|M.bovis_AF2122/97 RKLSRRYRVPVLSVHAPCLLISQRVWGANPILKLDRSVRAAEQLGAQTVV
Rv0498|M.tuberculosis_H37Rv RKLSRRYRVPVLSVHAPCLLISQRVWGANPILKLDRSVRAAEQLGAQTVV
MLBr_02432|M.leprae_Br4923 KGLSRRYRVPVLSVHAPCLLISQRVWGANPVPKLERSVRAAEQLGAQTVV
MAV_4654|M.avium_104 KKLSRRYRVPVLSVHAPCLLISQRVWGANPIPKLDRSVRAAEQLGAQTVV
Mflv_0077|M.gilvum_PYR-GCK AAMSKRYGVPVLSVHAPCLLITQRVWGANPITKLARSVRAAEQLGAQTVV
TH_0883|M.thermoresistible__bu ARLSKRYGVPVLSVHAPCLLITQRVWGANPVAKLERSVRAAEKLGAQTVV
Mvan_0835|M.vanbaalenii_PYR-1 GQMSEQYDVPVLSVHAPCLLITQRVWGANPVPKLERSVRAAEKLGAQTVV
MSMEG_0941|M.smegmatis_MC2_155 EKLSANYGVPVLSVHAPCLLISQRVWGANPITKLERSVCAAEQLGAQTVV
MAB_4011c|M.abscessus_ATCC_199 AKLSRKYGMPVLSVHAPCLLVSQRVWGANPIPKLERSVRAAEKLGAQTVV
:* .* :***********::********: ** *** ***:*******
MUL_4568|M.ulcerans_Agy99 VHPPFRWQRRYAEGFSDEVAALEASSDVVVAVENMFPFRADRFFGAGQ-S
MMAR_0824|M.marinum_M VHPPFRWQRRYAEGFSDQVAALEASSDVVVAVENMFPFRADRFFGAGQ-S
Mb0509|M.bovis_AF2122/97 VHPPFRWQRRYAEGFSDQVAALEAASTVMVAVENMFPFRADRFFGAGQ-S
Rv0498|M.tuberculosis_H37Rv VHPPFRWQRRYAEGFSDQVAALEAASTVMVAVENMFPFRADRFFGAGQ-S
MLBr_02432|M.leprae_Br4923 VHPPFRWQRRYAEGFSDQVAELEAASDVMIAVENMFPFRADRFFGADQ-S
MAV_4654|M.avium_104 VHPPFRWQRRYAEGFTEQVATLEAASDVMIAVENMFPFRADRFFGAGQ-S
Mflv_0077|M.gilvum_PYR-GCK VHPPFRWQRRYAEGFSDQVAELESGSDVLVAVENMFPFRADRFFGAGPTS
TH_0883|M.thermoresistible__bu VHPPFRWQRRYAEGFTDQVAELEASSDVLVAVENMFPFRADRFFGAGEAS
Mvan_0835|M.vanbaalenii_PYR-1 VHPPFRWQRRYAEGFSAQVAALEATSDVMVAVENMFPFRADRFFGAGQTS
MSMEG_0941|M.smegmatis_MC2_155 VHPPFRWQRRYAEGFSEQVAELEAGSDVMVAVENMFPFRADRFFGAGQTS
MAB_4011c|M.abscessus_ATCC_199 VHPPFRWQRRYAEGFAEQVAALEDSSDVHVAVENMFPLRADRLFGAGQTS
***************: :** ** * * :*******:****:***. *
MUL_4568|M.ulcerans_Agy99 RERMRKRGGGPGAAISAFAPSYDPLDGDHAHYTLDLSHTSTAGTDALSMA
MMAR_0824|M.marinum_M RERMRKRGGGPGAAISAFAPSYDPLDGDHAHYTLDLSHTSTAGTDALSMA
Mb0509|M.bovis_AF2122/97 RERMRKRGGGPGPAISAFAPSYDPLDGNHAHYTLDLSHTATAGTDSLDMA
Rv0498|M.tuberculosis_H37Rv RERMRKRGGGPGPAISAFAPSYDPLDGNHAHYTLDLSHTATAGTDSLDMA
MLBr_02432|M.leprae_Br4923 RERMRKRGGGPGPAISVFAPSFDPLAGNHAHYTLDLSHTATAGSDSLEMV
MAV_4654|M.avium_104 LERMRRRGGGPGPAISAFAPSYDPLDGHHAHYTLDLSHTATAGTDSLDMA
Mflv_0077|M.gilvum_PYR-GCK IERMRKRGGTPGPGISAFAPSYDPLDGGHAHYTLDLSHTATAGTDALDMA
TH_0883|M.thermoresistible__bu IERMRKRGGKPGPGISAFAPSYDPLDGNHAHYTLDLSHTATAGTDAMDMA
Mvan_0835|M.vanbaalenii_PYR-1 IERMRKRGGNPGPGISAFAPSYDPLDGGHAHYTLDLSHTATAGTDAVDMA
MSMEG_0941|M.smegmatis_MC2_155 IERMRKRGGRPGPGISAFAPSYDPLDGNHAHYTLDLSHTATAGTDAVDMA
MAB_4011c|M.abscessus_ATCC_199 VERMKRRGGRPGIGVSAFSPSFDPTDANHAHYTLDLSHTATAGTDALDMM
***::*** ** .:*.*:**:** . ***********:***:*::.*
MUL_4568|M.ulcerans_Agy99 RRMGSGLMHLHLCDGSGLPADEHLVPGRGTEPTAEVCQMLAGRGFAGQVV
MMAR_0824|M.marinum_M RRMGSGLMHLHLCDGSGLPADEHLVPGRGTEPTAEVCQMLAGGGFAGQVV
Mb0509|M.bovis_AF2122/97 RRMGPGLVHLHLCDGSGLPADEHLVPGRGTQPTAEVCQMLAGSGFVGHVV
Rv0498|M.tuberculosis_H37Rv RRMGPGLVHLHLCDGSGLPADEHLVPGRGTQPTAEVCQMLAGSGFVGHVV
MLBr_02432|M.leprae_Br4923 RRMGSGLVHLHLCDGSGLPADEHLVPGRGTQPTAAVCQLLAGADFAGHVV
MAV_4654|M.avium_104 RRMGNGLVHLHLCDGNGLPADEHLVPGRGTQPTAEVCQMLAAGHFAGHVI
Mflv_0077|M.gilvum_PYR-GCK ARMGEGLAHLHLCDGSGASYDEHLVPGRGTQPTAEVCEMLAASDFAGHVI
TH_0883|M.thermoresistible__bu RRMGDGLAHLHLCDGSGASVDEHLVPGRGTQPAAEICQMLAAGDFTGHVI
Mvan_0835|M.vanbaalenii_PYR-1 RRMGDGLVHLHLADGSGASTDEHLVPGRGTQPTVEVCEMLAASDFAGHVI
MSMEG_0941|M.smegmatis_MC2_155 RRMGDGLVHLHLADGNGASTDEHLVPGRGTQPVAQVCRMLAESDFAGHVV
MAB_4011c|M.abscessus_ATCC_199 RRMGSGLTHLHLTDGTGASVDEHLVPGKGTQPVAQVCQELAGGDFTGQVV
*** ** **** **.* . *******:**:*.. :*. ** *.*:*:
MUL_4568|M.ulcerans_Agy99 LEVSTSSARSAAEREAMLSESLQFARTHLMR
MMAR_0824|M.marinum_M LEVSTSSARSAAEREAMLSESLQFARTHLMR
Mb0509|M.bovis_AF2122/97 LEVSTSSARSANERESMLAESLQFARTHLLR
Rv0498|M.tuberculosis_H37Rv LEVSTSSARSANERESMLAESLQFARTHLLR
MLBr_02432|M.leprae_Br4923 LEVSTSSVRSATERETMLTESLQFARTYLLR
MAV_4654|M.avium_104 LEVSTSSARSATEREAMLAESLQFARTHLLR
Mflv_0077|M.gilvum_PYR-GCK LEVTTSGARSGAEREALLTESLQFARTHLLR
TH_0883|M.thermoresistible__bu LEVTTSRARNADEREALLTESLQFARQHLLR
Mvan_0835|M.vanbaalenii_PYR-1 LEVSTSGARTAAEREALLTESLQFARRHLLR
MSMEG_0941|M.smegmatis_MC2_155 LEVTTQTARTNGEREALLTESLQFARKHLLR
MAB_4011c|M.abscessus_ATCC_199 LEVHTSTARTEDQREAILRESLEFARTHLVR
*** *. .*. :**::* ***:*** :*:*