For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLAKVRDGVRFCAVCGNVSDDERCRIC ADPRRDGALVCVVEEPKDIQAVERTREYRGRYHVLGGAFDPLSGIGPEQLRIRELLTRIGDRVDGVDITE VIIATDPNTEGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALTGRRAMV*
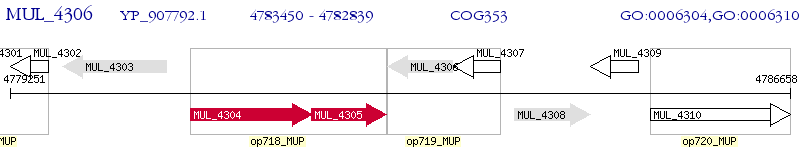
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4306 | recR | - | 100% (203) | recombination protein RecR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3742c | recR | 1e-105 | 90.59% (202) | recombination protein RecR |
| M. gilvum PYR-GCK | Mflv_1288 | recR | 1e-105 | 90.10% (202) | recombination protein RecR |
| M. tuberculosis H37Rv | Rv3715c | recR | 1e-105 | 90.59% (202) | recombination protein RecR |
| M. leprae Br4923 | MLBr_02329 | recR | 1e-104 | 90.15% (203) | recombination protein RecR |
| M. abscessus ATCC 19977 | MAB_0320 | recR | 1e-101 | 87.13% (202) | recombination protein RecR |
| M. marinum M | MMAR_5231 | recR | 1e-113 | 99.01% (203) | recombination protein RecR |
| M. avium 104 | MAV_0388 | recR | 1e-108 | 93.10% (203) | recombination protein RecR |
| M. smegmatis MC2 155 | MSMEG_6279 | recR | 1e-107 | 91.13% (203) | recombination protein RecR |
| M. thermoresistible (build 8) | TH_2287 | recR | 1e-104 | 90.10% (202) | PROBABLE RECOMBINATION PROTEIN RECR |
| M. vanbaalenii PYR-1 | Mvan_5527 | recR | 1e-105 | 90.59% (202) | recombination protein RecR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1288|M.gilvum_PYR-GCK MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLNRVR
Mvan_5527|M.vanbaalenii_PYR-1 MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLNKVR
MSMEG_6279|M.smegmatis_MC2_155 MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLGRIR
TH_2287|M.thermoresistible__bu VFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPEIDRLTAALNRVR
MUL_4306|M.ulcerans_Agy99 MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLAKVR
MMAR_5231|M.marinum_M MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLAKVR
Mb3742c|M.bovis_AF2122/97 MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPSDIDRLTGVLAKVR
Rv3715c|M.tuberculosis_H37Rv MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPSDIDRLTGVLAKVR
MAV_0388|M.avium_104 MFEGPVQDLIDELGKLPGIGPKSAQRIAFHLLSVEPPDIDRLTAVLARVR
MLBr_02329|M.leprae_Br4923 MFEGPVQNLIDELGKLPGIGPKSAQRIAFHLLSVEPQDIDRLTAVLVKVR
MAB_0320|M.abscessus_ATCC_1997 MFEGPVQDLIDELGKLPGVGPKSAQRIAFHLLGVEAPDIDRLTAALTRVR
:******:**********:*************.**. :*****..* ::*
Mflv_1288|M.gilvum_PYR-GCK DGVTFCSVCGNVSDEERCRICGDARRDASLICVVEEPKDVQAVERTREFR
Mvan_5527|M.vanbaalenii_PYR-1 DGVTFCAVCGNVSDEERCRICGDARRDASLICVVEEPKDVQAVERTREFR
MSMEG_6279|M.smegmatis_MC2_155 DGVTFCSVCGNVSDEERCRICSDPRRDASLVCVVEEPKDVQAVERTREFR
TH_2287|M.thermoresistible__bu DGVTFCEVCGNVSDAQRCRICSDSRRDASLVCVVEEPKDVQAVERTREFR
MUL_4306|M.ulcerans_Agy99 DGVRFCAVCGNVSDDERCRICADPRRDGALVCVVEEPKDIQAVERTREYR
MMAR_5231|M.marinum_M DGVRFCAVCGNVSDDERCRICADPRRDGSLVCVVEEPKDIQAVERTREYR
Mb3742c|M.bovis_AF2122/97 DGVRFCAVCGNVSDNERCRICSDIRRDASVVCIVEEPKDIQAVERTREFR
Rv3715c|M.tuberculosis_H37Rv DGVRFCAVCGNVSDNERCRICSDIRRDASVVCIVEEPKDIQAVERTREFR
MAV_0388|M.avium_104 DGVRFCAVCGNVSDDERCRICSDPRRDASVVCVVEEPKDVQAVERTREFR
MLBr_02329|M.leprae_Br4923 DDVRFCTVCGNVSDDERCRICSDTRRDASVVCVVEEPKDVQAVERTREFR
MAB_0320|M.abscessus_ATCC_1997 DGVQFCEVCGNVSDKERCRICADPRRDIALVCVVEEPKDVQAVERTREFR
*.* ** ******* :*****.* *** :::*:******:********:*
Mflv_1288|M.gilvum_PYR-GCK GRYHVLGGALDPLSGIGPDQLRIRELLNRIGERVDGVDVAEVIIATDPNT
Mvan_5527|M.vanbaalenii_PYR-1 GRYHVLGGALDPLSGIGPDQLRIRELLNRIGERVDGVEVAEVIIATDPNT
MSMEG_6279|M.smegmatis_MC2_155 GRYHVLGGALDPLSGIGPEQLRIRELLNRIGERVEGVDVAEVIIATDPNT
TH_2287|M.thermoresistible__bu GRYHVLGGALDPLSGIGPEQLRIRELLNRIGERVDGVDITEVIIATDPNT
MUL_4306|M.ulcerans_Agy99 GRYHVLGGAFDPLSGIGPEQLRIRELLTRIGDRVDGVDITEVIIATDPNT
MMAR_5231|M.marinum_M GRYHVLGGALDPLSGIGPEQLRIRELLTRIGDRVDGVDITEVIIATDPNT
Mb3742c|M.bovis_AF2122/97 GRYHVLGGALDPLSGIGPDQLRIRELLSRIGERVDDVDVTEVIIATDPNT
Rv3715c|M.tuberculosis_H37Rv GRYHVLGGALDPLSGIGPDQLRIRELLSRIGERVDDVDVTEVIIATDPNT
MAV_0388|M.avium_104 GRYHVLGGALDPLSGVGPDQLRIRELLSRIGERVDDVDITEVIIATDPNT
MLBr_02329|M.leprae_Br4923 GRYHVLGGALDPLSGIGPEQLRIRELLSRIGERVDDVGITEVIIATDPNT
MAB_0320|M.abscessus_ATCC_1997 GRYHVLGGALDPLSGVGPDQLRIRELLTRIGTEEDGVSISEVIIATDPNT
*********:*****:**:********.*** . :.* ::**********
Mflv_1288|M.gilvum_PYR-GCK EGEATATYLVRMLRDIPGLSVTRIASGLPMGGDLEFADELTLGRALAGRR
Mvan_5527|M.vanbaalenii_PYR-1 EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALAGRR
MSMEG_6279|M.smegmatis_MC2_155 EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALAGRR
TH_2287|M.thermoresistible__bu EGEATATYLVRILRDIPGLTVTRIASGLPMGGDLEFADELTLGRALVGRR
MUL_4306|M.ulcerans_Agy99 EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALTGRR
MMAR_5231|M.marinum_M EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALTGRR
Mb3742c|M.bovis_AF2122/97 EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALAGRR
Rv3715c|M.tuberculosis_H37Rv EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALAGRR
MAV_0388|M.avium_104 EGEATATYLVRMLRDIPGLTVTRIASGLPMGGDLEFADELTLGRALAGRR
MLBr_02329|M.leprae_Br4923 EGEATATYLVRMLRDMPGLTVTRIASGLPMGGDLEFADELTLGRALTGRR
MAB_0320|M.abscessus_ATCC_1997 EGEATATYLVRMLRDFPGLTVTRLASGLPMGGDLEFADELTLGRALSGRR
***********:***:***:***:********************** ***
Mflv_1288|M.gilvum_PYR-GCK AMA
Mvan_5527|M.vanbaalenii_PYR-1 AMA
MSMEG_6279|M.smegmatis_MC2_155 AMV
TH_2287|M.thermoresistible__bu AMT
MUL_4306|M.ulcerans_Agy99 AMV
MMAR_5231|M.marinum_M AMV
Mb3742c|M.bovis_AF2122/97 VLA
Rv3715c|M.tuberculosis_H37Rv VLA
MAV_0388|M.avium_104 AMV
MLBr_02329|M.leprae_Br4923 TMV
MAB_0320|M.abscessus_ATCC_1997 PL-
: