For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGGRGSREQLAVPRAQFRSYYGRPVLKTPAWEWKIAAYLFSGGLSAGSALLAAGADLNGQPALRRVSRV GALASIATGSYFLIADLGRPERFHHMLRVAKPSSPMSVGTWILAAYGPGAGLAGAAELMPARLRRTWLGA LLARLARPAGLSAAAVAPGVASYTAVLLSQTAVPAWHEAHPYLPFVFTGSAAASGGGLGMLLAPVAEAAP ARRMSVAGAALEVAASRLLERRLGLVGEAYTTGPAHRTRKWAEYLTVGGALGTLAAGRNRLGAALCGLAL LTGSALQRFGVFEAGVESTRDPKYVVVPQRERVDTGRAARGGS
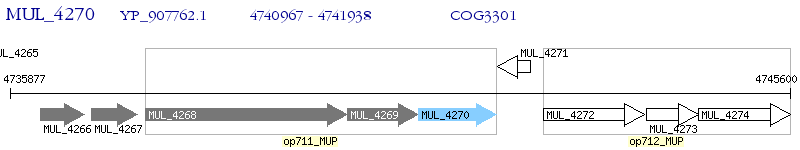
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4270 | nrfD | - | 100% (323) | formate-dependent nitrite reductase, membrane component, NrfD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_5197 | nrfD | 1e-180 | 98.45% (323) | formate-dependent nitrite reductase, membrane component, |
| M. avium 104 | MAV_0424 | - | 6e-80 | 50.00% (320) | polysulphide reductase, NrfD superfamily protein |
| M. smegmatis MC2 155 | MSMEG_1846 | - | 1e-140 | 77.60% (317) | polysulphide reductase, NrfD |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4270|M.ulcerans_Agy99 -----------------------MTGGR----GSREQLAVPRAQFRSYYG
MMAR_5197|M.marinum_M -----------------------MTGGR----GSREQLAVPRAQFRSYYG
MSMEG_1846|M.smegmatis_MC2_155 -------MRELPSDAGAADMHAQTRHRR----GGRDDMAVPPAEFRSYYG
MAV_0424|M.avium_104 MSTSEFDSFRPPEPAGGKRRKGKRGGRRGAGDGSREMPMVPEAEFSSYYG
* *.*: ** *:* ****
MUL_4270|M.ulcerans_Agy99 RPVLKTPAWEWKIAAYLFSGGLSAGSALLAAGADLNGQPALRRVSRVGAL
MMAR_5197|M.marinum_M RPVLKTPAWEWKIAAYLFSGGLSAGSALLAAGADLNGQPALRRVSRVGAL
MSMEG_1846|M.smegmatis_MC2_155 RQILKTPVWNWMIAAYLFSGGLSAGSALLAAGADLTGRPQLRKVSRVGAL
MAV_0424|M.avium_104 RPVVKPAPWGHEVGAYLFLGGVAGGSGLLAAGAQLTGRDTLRRNTRLTAL
* ::*.. * :.**** **::.**.******:*.*: **: :*: **
MUL_4270|M.ulcerans_Agy99 ASIATGSYFLIADLGRPERFHHMLRVAKPSSPMSVGTWILAAYGPGAGLA
MMAR_5197|M.marinum_M ASIAAGSYFLIADLGRPQRFHHMLRVAKPSSPMSVGTWILAAYGPGAGLA
MSMEG_1846|M.smegmatis_MC2_155 VSLLASMYFLVGDLGRPERFHHMLRVAKPSSPMSMGTWILSGYGPGAGAA
MAV_0424|M.avium_104 AAVLLSLVALVKDLGRPERFLNMLRTVKLTSPMNLGSWILSAFSAGISVA
.:: . *: *****:** :***..* :***.:*:***:.:..* . *
MUL_4270|M.ulcerans_Agy99 GAAELMPARLRRTWLG---ALLARLARPAGLSAAAVAPGVASYTAVLLSQ
MMAR_5197|M.marinum_M GAAELMPARLRRTWPG---ALLARLARPAGLSAAAVAPGVASYTAVLLSQ
MSMEG_1846|M.smegmatis_MC2_155 AVAELMPKRLRRSRLG---RLLDFAARPAGLEAAAVAPGVASYTAVLLSQ
MAV_0424|M.avium_104 AAAEVDRMTGQRLPLGPLRPVLRAAEGPAGLQAAVLAPPLAVYTAVLLAD
..**: :* * :* ****.**.:** :* ******::
MUL_4270|M.ulcerans_Agy99 TAVPAWHEAHPYLPFVFTGSAAASGGGLGMLLAPVAEAAPARRMSVAGAA
MMAR_5197|M.marinum_M TAVPAWHEAHPYLPFVFTGSAAASGGGLGMLLAPVAEAAPARRMSVAGAV
MSMEG_1846|M.smegmatis_MC2_155 TAVPAWREAHPYLPFIFTGSAAASGAGLGMLLAPVSESGPARRMAVAGAA
MAV_0424|M.avium_104 TATPTWNAAHEDLPFVFVSSASLAASGLAMVTTPVHQAGPARTLAVLGAL
**.*:*. ** ***:*..**: :..**.*: :** ::.*** ::* **
MUL_4270|M.ulcerans_Agy99 LEVAASRLLERRLG-LVGEAYTTGPAHRTRKWAEYLTVGGALGTLAAGRN
MMAR_5197|M.marinum_M LEVAASRLLERRLG-LVGEAYTTGPAHRTRKWAEYLTVGGALGTLAAGRN
MSMEG_1846|M.smegmatis_MC2_155 LEVAASRTMEHRLG-LVGEAYTTGRPHTLRKWSEILTVGGAVGAVASGRN
MAV_0424|M.avium_104 GDLAASKVMERRMDPVAAEPLHTGGPGRMLRASERLVIAGGLGTLLGGRH
::***: :*:*:. :..*. ** . : :* *.:.*.:*:: .**:
MUL_4270|M.ulcerans_Agy99 RLGAALCGLALLTGSALQRFGVFEAGVESTRDPKYVVVPQRERVDTGRAA
MMAR_5197|M.marinum_M RLGAALCGLALLTGSALQRFGVFEAGVESTRDPKYVVVPQRERVDTGRSA
MSMEG_1846|M.smegmatis_MC2_155 RWAVAASGAALLAGSALQRFGVFEAGVQSTQDPKYVVIPQRERLDAGHPA
MAV_0424|M.avium_104 RAVAVVSGLALATASALTRFGVFEAGLESARHPRYTIEPQQRRLAARRAA
* .. .* ** :.*** ********::*::.*:*.: **:.*: : :.*
MUL_4270|M.ulcerans_Agy99 RGGS-------
MMAR_5197|M.marinum_M RGGS-------
MSMEG_1846|M.smegmatis_MC2_155 RGDDR------
MAV_0424|M.avium_104 GITSDSITTAG
.