For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VADVELYLDPICPFAWVTSRWLLEAAQSTHTPVTLRQMSLAVLNEGKDLDAKQQQMMQRSRQLGRLFAAV TTKYGADAFARLYDSIGTRIRLRREEITQGGIREALAETGLDESLSETLDDSGLDDEARHAHQASQDALG GTAGSPIIAVDGQGFFGPVLTRMPSGADGVRLLEAVLTAAKTPEFAVLQRPYQGPPVLDGTAR
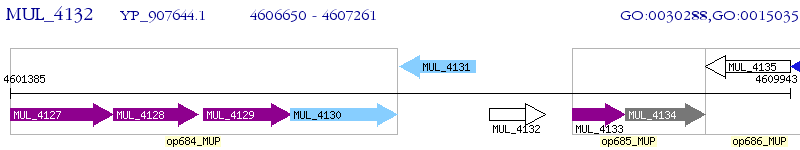
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4132 | - | - | 100% (203) | hypothetical protein MUL_4132 |
| M. ulcerans Agy99 | MUL_2708 | - | 1e-16 | 29.47% (190) | hypothetical protein MUL_2708 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2493c | - | 2e-31 | 35.71% (196) | hypothetical protein Mb2493c |
| M. gilvum PYR-GCK | Mflv_2578 | - | 7e-30 | 36.50% (200) | DsbA oxidoreductase |
| M. tuberculosis H37Rv | Rv2466c | - | 2e-31 | 35.71% (196) | hypothetical protein Rv2466c |
| M. leprae Br4923 | MLBr_01485 | - | 4e-31 | 35.23% (193) | hypothetical protein MLBr_01485 |
| M. abscessus ATCC 19977 | MAB_4744c | - | 3e-34 | 42.19% (192) | hypothetical protein MAB_4744c |
| M. marinum M | MMAR_5056 | - | 1e-112 | 99.51% (203) | hypothetical protein MMAR_5056 |
| M. avium 104 | MAV_1706 | - | 9e-33 | 36.73% (196) | hypothetical protein MAV_1706 |
| M. smegmatis MC2 155 | MSMEG_4688 | - | 1e-33 | 37.82% (193) | hypothetical protein MSMEG_4688 |
| M. thermoresistible (build 8) | TH_0346 | - | 3e-33 | 36.41% (195) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4056 | - | 1e-32 | 38.19% (199) | DsbA oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2578|M.gilvum_PYR-GCK --------MSEKSRADFWFDPLCPWAWITSRWVLEAEKVRDIEVNFHVMS
Mvan_4056|M.vanbaalenii_PYR-1 --------MSEKSRTDFWFDPLCPWAWITSRWVLEVEKVRDIEVNFHVMS
MSMEG_4688|M.smegmatis_MC2_155 --------MAQKDVAGFWFDPLCPWCWITSRWILEVEKVRDIEVQFHVMS
MAV_1706|M.avium_104 ----MAQQAPAKNKADFWFDPLCPWAWITSRWILEVEKVRDIEVNFHVMS
TH_0346|M.thermoresistible__bu --------MAAKDTADFWFDPLCPWAWITSRWILEVEKVRDIDVNFRVMS
Mb2493c|M.bovis_AF2122/97 ----MLEKAPQKSVADFWFDPLCPWCWITSRWILEVAKVRDIEVNFHVMS
Rv2466c|M.tuberculosis_H37Rv ----MLEKAPQKSVADFWFDPLCPWCWITSRWILEVAKVRDIEVNFHVMS
MLBr_01485|M.leprae_Br4923 ----MPEKASVKYQADLWFDPLCPWCWITSRWILEVEKVRDVEVHFHVMS
MUL_4132|M.ulcerans_Agy99 -----------MADVELYLDPICPFAWVTSRWLLEAAQSTHTPVTLRQMS
MMAR_5056|M.marinum_M -----------MADVELYLDPICPFAWVTSRWLLEAAQSTHTPVTLRQMS
MAB_4744c|M.abscessus_ATCC_199 MPADGDAAASGPFDVRIWLDPVCPFSWNTARWLDAVAIGSGIPVDWQLMN
. :::**:**:.* *:**: . * : *.
Mflv_2578|M.gilvum_PYR-GCK LAVLNEGRD-LPEEYLEMMKTAWGPVRVAIAAEQAKGPEVLGPLYTAFGT
Mvan_4056|M.vanbaalenii_PYR-1 LAVLNEGKD-VPEAYREAMQKAWGPVRVAIAAEQAKGPEILGPLYTAMGT
MSMEG_4688|M.smegmatis_MC2_155 LAVLNEGRD-LPEEYQEMMKKAWGPVRVAIAAEQLKGSEILSPLYTAMGT
MAV_1706|M.avium_104 LAVLNENREGLPDEYRERLAKAFGPVRVAIAAEQAHGAEVLAPLYTAMGT
TH_0346|M.thermoresistible__bu LAVLNEGRD-VPDEYRETMKKAWAPVRVAIAAEQAHGREVLSPLYTAMGT
Mb2493c|M.bovis_AF2122/97 LAILNENRDDLPEQYREGMARAWGPVRVAIAAEQAHGAKVLDPLYTAMGN
Rv2466c|M.tuberculosis_H37Rv LAILNENRDDLPEQYREGMARAWGPVRVAIAAEQAHGAKVLDPLYTAMGN
MLBr_01485|M.leprae_Br4923 LAILNENREDLTENYLENITKAWGPARVVIAAEQANGASVLDPLYTAMGI
MUL_4132|M.ulcerans_Agy99 LAVLNEGKD-LDAKQQQMMQRSRQLGRLFAAVTTKYGADAFARLYDSIGT
MMAR_5056|M.marinum_M LAVLNEGKD-LDAKQQQMMQRSRQLGRLFAAVTTKYGADAFARLYDSIGT
MAB_4744c|M.abscessus_ATCC_199 LAVLNEGRE-LPDVQQARMRDSRRVGRLMAGIQCELGQQGLRTAYFAFGR
**:***.:: : : : *: . * . : * ::*
Mflv_2578|M.gilvum_PYR-GCK RIHNLGYRETDPDFTRVIAESLDEVGLPAELAAAATSTDYDEALRKSHHE
Mvan_4056|M.vanbaalenii_PYR-1 RIHNQGYQGSDPDFSRVIRESLDEVGLPAELAEAATSTFYDVALRESHHA
MSMEG_4688|M.smegmatis_MC2_155 RIHNQ----DNKNFDEVIAQSLEEVGLPAELAEAATTDKYDEALRRSHHA
MAV_1706|M.avium_104 RIHNEG----NKDLTDVIKQSLAEVGLPAELAEAADTDAYDDALRKSHHA
TH_0346|M.thermoresistible__bu RIHNQG----NKDLDEVIAQSLAEVGLPAELAEAATTDRYDEALRKSHHE
Mb2493c|M.bovis_AF2122/97 RIHNQG----NHELDEVITQSLADAGLPAELAKAATSDAYDNALRKSHHA
Rv2466c|M.tuberculosis_H37Rv RIHNQG----NHELDEVITQSLADAGLPAELAKAATSDAYDNALRKSHHA
MLBr_01485|M.leprae_Br4923 RIHNED----NKNLDEVIKRSLADTGLPAELAAAAQSNAYDDALRESHHA
MUL_4132|M.ulcerans_Agy99 RIRLRR----EEITQGGIREALAETGLDESLSETLDDSGLDDEARHAHQA
MMAR_5056|M.marinum_M RIHLRR----EEITQGGIREALAETGLDESLSETLDDSGLDDEARHAHQA
MAB_4744c|M.abscessus_ATCC_199 LYFDES----LPLDAELADRVLGAVAPLTTTRETLSDASLDAEVRRSHEA
. * .. : * *.:*.
Mflv_2578|M.gilvum_PYR-GCK GMDPVGPDVGTPTIHVNGVAFFGPVLSRIPRGEEAGKLWDASVIFASYPH
Mvan_4056|M.vanbaalenii_PYR-1 GMDAVGDDVGTPTIHVNGVAFFGPVLSRIPRGEDAGKLWDASVIFASYPH
MSMEG_4688|M.smegmatis_MC2_155 GMDAVGDDVGTPTIHVNGVAFFGPVLSRIPRGEEAGKLWDASVTFASYPH
MAV_1706|M.avium_104 GMDPVGDDVGTPTIHVNNVAFFGPVLSRIPRGEEAGKLWDASVTFASYPH
TH_0346|M.thermoresistible__bu GMDAVGEDVGTPTIHVNNVAFFGPVLSRIPRGEEAGKLWDASVTFASYPH
Mb2493c|M.bovis_AF2122/97 GMDAVGEDVGTPTIHVNGVAFFGPVLSKIPRGEEAGKLWDASVTFASYPH
Rv2466c|M.tuberculosis_H37Rv GMDAVGEDVGTPTIHVNGVAFFGPVLSKIPRGEEAGKLWDASVTFASYPH
MLBr_01485|M.leprae_Br4923 GMDPVGDDVGTPTVHVNGVAFFGPVLSKIPRGEEAGKLWDASLTLAAYPH
MUL_4132|M.ulcerans_Agy99 SQDALGGTAGSPIIAVDGQGFFGPVLTRMPSGADGVRLLEAVLTAAKTPE
MMAR_5056|M.marinum_M SQDALGGTAGSPIIAVDGQGFFGPVLTRMPSGADGVRLLEAVLTAAKTPE
MAB_4744c|M.abscessus_ATCC_199 GQRALGDVGGSPLMQVNGRTSFGPVLTAVPEHDSARALFDAVAALARVPE
. .:* *:* : *:. *****: :* .. * :* * *.
Mflv_2578|M.gilvum_PYR-GCK FWELKRTRTESPEFD----
Mvan_4056|M.vanbaalenii_PYR-1 FWELKRSRTEAPVFD----
MSMEG_4688|M.smegmatis_MC2_155 FWELKRSRTESPEFD----
MAV_1706|M.avium_104 FFELKRTRDEKPEFD----
TH_0346|M.thermoresistible__bu FWELKRTRTEAPEFD----
Mb2493c|M.bovis_AF2122/97 FFELKRTRTEPPQFD----
Rv2466c|M.tuberculosis_H37Rv FFELKRTRTEPPQFD----
MLBr_01485|M.leprae_Br4923 FFELKRTRTEPPQFA----
MUL_4132|M.ulcerans_Agy99 FAVLQRPYQGPPVLDGTAR
MMAR_5056|M.marinum_M FAVLQRPYQGPPVLDGTAR
MAB_4744c|M.abscessus_ATCC_199 FAQLQRPRPAH--------
* *:*.