For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRTDQLAGTHLAPEPNDLALLVAGSHHNPHGVLGAHEYGDHIVIRAFRPHAAEVIAIVGDDRFPMQHIE SGLFAVALPFVDLIDYRLQVSYENSEPFTVADAYRFLPTLGEVDLHLFAEGRHERLWEALGAHPRSFTTA DGVVHGVSFAVWAPNAEGVSLIGDFNGWSGSEAPMRVLGSSGVWELFWPDFPADGLYKFRVHGADGVVSE RADPFAFGTEVPPQTASRVTVSDYSWGDGDWMTERAQRNPVFEPMSTYEVHLGSWRPGLSYRQLATELTD YVVAHKFTHVELLPVAEHPFAGSWGYQVTSYYAPTSRFGTPDDFRALVDALHQAGIGVIVDWVPAHFPKD AWALGRFDGTPLYEHSDPKRGEQLDWGTYVFDFGRPEVRNFLVANALYWIEQFHIDGLRVDAVASMLYLD YSRPQDGWTPNIYEGRENLEAVQFLQEMNATAHKAAPGIVTIAEESTSWPGVTRPTSIGGLGFSMKWNMG WMHDTLDYVSRDPIYRSYHHHEMTFSMLYAFSENYVLPLSHDEVVHGKGTLWGRMPGSNHTKAAGLRSLL AYQWAHPGKQLLFMGQEFGQRAEWSEQRGLDWFQLDENGFSNGVLRLVRDINEIYCDHCALWSQDTTPEG YSWIDANDSANNVLSFLRHGSDGSVMACIFNFAGSEHSDYRLGLPIAGRWREVLNTDATIYNGSGVGNFG GVDATAEPWHGRPASAVLVLPPSSALWLEPA
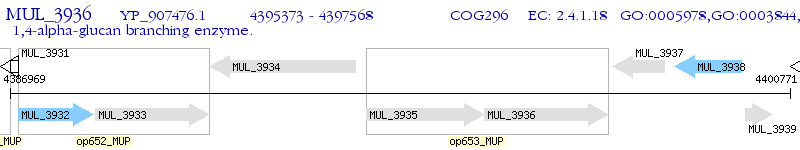
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3936 | glgB | - | 100% (731) | glycogen branching enzyme |
| M. ulcerans Agy99 | MUL_1553 | treZ | 2e-19 | 27.83% (345) | maltooligosyltrehalose trehalohydrolase, TreZ |
| M. ulcerans Agy99 | MUL_1555 | treX | 4e-10 | 24.72% (356) | maltooligosyltrehalose synthase TreX |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1361c | glgB | 0.0 | 88.78% (731) | glycogen branching enzyme |
| M. gilvum PYR-GCK | Mflv_2343 | - | 0.0 | 77.37% (729) | glycogen branching enzyme |
| M. tuberculosis H37Rv | Rv1326c | glgB | 0.0 | 88.37% (731) | glycogen branching enzyme |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1467c | - | 0.0 | 77.35% (724) | glycogen branching enzyme |
| M. marinum M | MMAR_4072 | glgB | 0.0 | 99.32% (731) | 1,4-alpha-glucan branching enzyme GlgB |
| M. avium 104 | MAV_3212 | treZ | 2e-19 | 27.06% (377) | malto-oligosyltrehalose trehalohydrolase |
| M. smegmatis MC2 155 | MSMEG_4918 | glgB | 0.0 | 80.74% (732) | glycogen branching enzyme |
| M. thermoresistible (build 8) | TH_1050 | glgB | 0.0 | 78.34% (697) | PROBABLE 1,4-ALPHA-GLUCAN BRANCHING ENZYME GLGB (GLYCOGEN |
| M. vanbaalenii PYR-1 | Mvan_4302 | - | 0.0 | 79.42% (729) | glycogen branching enzyme |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2343|M.gilvum_PYR-GCK MTNASNLKNQIDSPHLRPHTADLNRLLAGEHHDPHSVLGAHEYD-----D
Mvan_4302|M.vanbaalenii_PYR-1 MTNVSNLNNQVDSPHLRPHTADLNRLLAGEHHDPHAILGAHEYD-----D
MUL_3936|M.ulcerans_Agy99 MSRT----DQLAGTHLAPEPNDLALLVAGSHHNPHGVLGAHEYG-----D
MMAR_4072|M.marinum_M MSRT----DQLAGTHLAPEPNDLARLVAGSHHNPHGVLGAHEYG-----D
Mb1361c|M.bovis_AF2122/97 MSRS----EKLTGEHLAPEPAEMARLVAGTHHNPHGILGAHEYG-----D
Rv1326c|M.tuberculosis_H37Rv MSRS----EKLTGEHLAPEPAEMARLVAGTHHNPHGILGAHEYD-----D
MSMEG_4918|M.smegmatis_MC2_155 MTRSS---NQLTDAHLRPDPSDIHRLLAGEHHDPHSVLGAHEYP-----G
TH_1050|M.thermoresistible__bu ----------VTSPHLRPDPAELGRLLAGEHHNPHSILGAHEYG-----D
MAB_1467c|M.abscessus_ATCC_199 --------MSPSTAPLQPDAADIARLLAGTHHNPHSILGAHEYPRSKSKG
MAV_3212|M.avium_104 MTEQK---------------------------------------------
Mflv_2343|M.gilvum_PYR-GCK HTVIRAYRPHAVSVEALIGGTRYPLRHIESGLFAVAVPFTGLIDYRLAIR
Mvan_4302|M.vanbaalenii_PYR-1 HTVIRAYRPHAVAVVAVVGKNRYPLRHIEAGLFAVAVPFTNLIDYRFEIR
MUL_3936|M.ulcerans_Agy99 HIVIRAFRPHAAEVIAIVGDDRFPMQHIESGLFAVALPFVDLIDYRLQVS
MMAR_4072|M.marinum_M HTVIRAFRPHAAEVIAIVGDDRFPMQHIESGLFAVALPFVDLIDYRLQVS
Mb1361c|M.bovis_AF2122/97 HTVIRAFRPHAVEVVALVGKDRFSLQHLDSGLFAVALPFVDLIDYRLQVT
Rv1326c|M.tuberculosis_H37Rv HTVIRAFRPHAVEVVALVGKDRFSLQHLDSGLFAVALPFVDLIDYRLQVT
MSMEG_4918|M.smegmatis_MC2_155 HTVIRAFRPHAVKVTAVIGGERHVMQHLESGLFAVAVPFTNLIDYRLEID
TH_1050|M.thermoresistible__bu HTVLRAYRPHAQSVVALVGSDRYPLEHLESGLFAVALPFTGLMDYRLEIT
MAB_1467c|M.abscessus_ATCC_199 YTVIRAFKPGASAVAAVVGDAIHHMTHLGSGLFAVALPFLNLLDYRLDVE
MAV_3212|M.avium_104 -TEFRVWAPKPARVRLDVEGRPHAMSRTDDGWWHAAVACAPDARYGFLLD
:*.: * . * : . : : * : .*:. * : :
Mflv_2343|M.gilvum_PYR-GCK YSDD-ESAFVHTVADAYRFLPTLGEIDLHLFGEGRHERLWEILGAHRRTF
Mvan_4302|M.vanbaalenii_PYR-1 YSED-ESAFVHTVADAYRFLPTLGEVDLHLFAEGRHERLWEILGAHPRSF
MUL_3936|M.ulcerans_Agy99 YENS----EPFTVADAYRFLPTLGEVDLHLFAEGRHERLWEALGAHPRSF
MMAR_4072|M.marinum_M YENS----EPFTVADAYRFLPTLGEVDLHLFAEGRHERLWEALGAHPRSF
Mb1361c|M.bovis_AF2122/97 YEGC----EPHTVADAYRFLPTLGEVDLHLFAEGRHERLWEVLGAHPRSF
Rv1326c|M.tuberculosis_H37Rv YEGC----EPHTVADAYRFLPTLGEVDLHLFAEGRHERLWEVLGAHPRSF
MSMEG_4918|M.smegmatis_MC2_155 YPGVGDAVVTHTTADAYRFLPTLGELDLHLFAEGRHERLWEILGAHRRTF
TH_1050|M.thermoresistible__bu YPHS----GTHVVADAYRFLPTLGELDLHLFNEGRHERLWEILGAHPRTF
MAB_1467c|M.abscessus_ATCC_199 YGGT-----SLVIADAYRFLPTLGEVDLHLFNEGRHERLWEILGAHPRSF
MAV_3212|M.avium_104 DDPT--------------------------------------VLPDPRSP
: .. *:
Mflv_2343|M.gilvum_PYR-GCK TTADGEVTGVSFAVWAPNAKGVSVTGDFNHWG-NEAQMRVLGSTGVWEVF
Mvan_4302|M.vanbaalenii_PYR-1 TTSDGVVNGVSFAVWAPNAKGVSVTGDFNHWG-NEAQMRVLGSTGVWELF
MUL_3936|M.ulcerans_Agy99 TTADGVVHGVSFAVWAPNAEGVSLIGDFNGWSGSEAPMRVLGSSGVWELF
MMAR_4072|M.marinum_M TTADGVVHGVSFAVWAPNAEGVSLIGDFNGWSGSEAPMRVLGSSGVWELF
Mb1361c|M.bovis_AF2122/97 TTADGVVSGVSFAVWAPNAKGVSLIGEFNGWNGHEAPMRVLGPSGVWELF
Rv1326c|M.tuberculosis_H37Rv TTADGVVSGVSFAVWAPNAKGVSLIGEFNGWNGHEAPMRVLGPSGVWELF
MSMEG_4918|M.smegmatis_MC2_155 TTADGPVSGVSFAVWAPNAKGVSLIGEFNHWGGNDAPMRVLGSSGVWELF
TH_1050|M.thermoresistible__bu RTADGEVSGVSFAVWAPNAKGVSLIGDFNGWSGTDAPMRVLGSSGVWEVF
MAB_1467c|M.abscessus_ATCC_199 VTADGEVSGVSFAVWAPNARGVSVIGDFNGWNGNDAPMRTLGSSGVWELF
MAV_3212|M.avium_104 RQPEGVHARS--QLWDP---------------------------------
.:* :* *
Mflv_2343|M.gilvum_PYR-GCK WPNFPDGGLYKFRVHGADGSVVDRADPMAFATEVPPQTASKVFESEYTWD
Mvan_4302|M.vanbaalenii_PYR-1 WPNFPDGGLYKFRVHGADGSVTDRADPMAFATEVPPQTASRVFRSDYTWS
MUL_3936|M.ulcerans_Agy99 WPDFPADGLYKFRVHGADGVVSERADPFAFGTEVPPQTASRVTVSDYSWG
MMAR_4072|M.marinum_M WPDFPADGLYKFRVHGADGVVSERADPFAFGTEVPPQTASRVTVSDYSWG
Mb1361c|M.bovis_AF2122/97 WPDFPCDGLYKFRVHGADGVVTDRADPFAFGTEVPPQTASRVTSSDYTWG
Rv1326c|M.tuberculosis_H37Rv WPDFPCDGLYKFRVHGADGVVTDRADPFAFGTEVPPQTASRVTSSDYTWG
MSMEG_4918|M.smegmatis_MC2_155 WPGFPPDGLYKFRVHGADGSVTDRADPMAFATEVPPHTASRVTESTYEWH
TH_1050|M.thermoresistible__bu WPDFPVGGLYKFRIHGADGSVTDRADPMAFATEVPPNTASRVTRSEYTWS
MAB_1467c|M.abscessus_ATCC_199 WPGMTLGALYKFRVHGADGSVTDRADPFAFATELPPATASRVSTTSYSWE
MAV_3212|M.avium_104 --------------------------------------------AAATWT
: *
Mflv_2343|M.gilvum_PYR-GCK DAEWMDGRTLRNPVFEPMSTYEVHLGSWRPGLSYTELAEQLTEYLVQHGF
Mvan_4302|M.vanbaalenii_PYR-1 DQEWMSGRALRNPVFEPMSTYEVHLGSWRPGLSYIELAEQLTDYVVEHGF
MUL_3936|M.ulcerans_Agy99 DGDWMTERAQRNPVFEPMSTYEVHLGSWRPGLSYRQLATELTDYVVAHKF
MMAR_4072|M.marinum_M DGDWMTERAQRNPVFEPMSTYEVHLGSWRPGLSYRQLATELTDYVVSHKF
Mb1361c|M.bovis_AF2122/97 DDDWMAGRALRNPVNEAMSTYEVHLGSWRPGLSYRQLARELTDYIVDQGF
Rv1326c|M.tuberculosis_H37Rv DDDWMAGRALRNPVNEAMSTYEVHLGSWRPGLSYRQLARELTDYIVDQGF
MSMEG_4918|M.smegmatis_MC2_155 DEDWMTRRASLNPVFEPMSTLEVHLLSWRPGLTYRQLATELTEYVVEQGF
TH_1050|M.thermoresistible__bu DDDWMARRALRNPVFEPMSTYEVHLMSWRPGLSYRELATELTEYVVEQGF
MAB_1467c|M.abscessus_ATCC_199 DGEWMSQRASRNPVFEPMSTYEVHLASWRPGLNYRDLARELTEYVLAQGF
MAV_3212|M.avium_104 DSAWPGRSTHG------AVIYELHLGTFTAAGTF-DSAIEKLDYLVDLGV
* * : *:** :: .. .: : * : :*:: .
Mflv_2343|M.gilvum_PYR-GCK THVEMLPVAEHPFGGSWGYQVTSYYAPSSRFGSPDEFRYLVDALHRAGIG
Mvan_4302|M.vanbaalenii_PYR-1 THVEMLPVAEHPFGGSWGYQVTSYYAPSSRFGTPDEFRYLVDALHRAGIG
MUL_3936|M.ulcerans_Agy99 THVELLPVAEHPFAGSWGYQVTSYYAPTSRFGTPDDFRALVDALHQAGIG
MMAR_4072|M.marinum_M THVELLPVAEHPFAGSWGYQVTSYYAPTSRFGTPDDFRALVDALHQAGIG
Mb1361c|M.bovis_AF2122/97 THVELLPVAEHPFAGSWGYQVTSYYAPTSRFGTPDDFRALVDALHQAGIG
Rv1326c|M.tuberculosis_H37Rv THVELLPVAEHPFAGSWGYQVTSYYAPTSRFGTPDDFRALVDALHQAGIG
MSMEG_4918|M.smegmatis_MC2_155 THVEMLPVAEHPFGGSWGYQVTSYYAPTSRLGTPDDFRYLVDALHQAGIG
TH_1050|M.thermoresistible__bu THVELLPVAEHPFGGSWGYQVTSYYAPTSRLGTPDDFRFLVDSLHQAGIG
MAB_1467c|M.abscessus_ATCC_199 THVELLPVAEHPFGGSWGYQVTSYYAPTSRLGTPDDFRYLVDALHRAGIG
MAV_3212|M.avium_104 DFVELMPVNSFAGTRGWGYDGVLWYSVHEPYGGPDGLVRFVDACHARGLG
.**::** ... .***: . :*: . * ** : :**: * *:*
Mflv_2343|M.gilvum_PYR-GCK VLVDWVPAHFPKDSWALGRFDGTALYEHGDPRRGEQLDWGTYVFDFGRAE
Mvan_4302|M.vanbaalenii_PYR-1 VIVDWVPAHFPKDAWALGRFDGTALYEHGDPRRGEQLDWGTYVFDFGRSE
MUL_3936|M.ulcerans_Agy99 VIVDWVPAHFPKDAWALGRFDGTPLYEHSDPKRGEQLDWGTYVFDFGRPE
MMAR_4072|M.marinum_M VIVDWVPAHFPKDAWALGRFDGTPLYEHSDPKRGEQLDWGTYVFDFGRPE
Mb1361c|M.bovis_AF2122/97 VIVDWVPAHFPKDAWALGRFDGTPLYEHSDPKRGEQLDWGTYVFDFGRPE
Rv1326c|M.tuberculosis_H37Rv VIVDWVPAHFPKDAWALGRFDGTPLYEHSDPKRGEQLDWGTYVFDFGRPE
MSMEG_4918|M.smegmatis_MC2_155 VIVDWVPAHFPKDAWALGRFDGTALYEHSDPRRGEQLDWGTYVFDFGRPE
TH_1050|M.thermoresistible__bu VIIDWVPAHFPKDAWALGRFDGTALYEHADPRRGEQLDWGTYVFDFGRPE
MAB_1467c|M.abscessus_ATCC_199 VLVDWVPAHFPKDAWALARFDGTPLYEHADPHRAEQLDWGTYVFDFGRPE
MAV_3212|M.avium_104 VLIDAVFNHLGPSGNYLPRFG-PYLSSASNP-----WGEGINIADADSDE
*::* * *: .. * **. . * . .:* . * : * . *
Mflv_2343|M.gilvum_PYR-GCK VRNFLVANALYWLQEYHIDGLRVDAVASMLY------LDYSRPEGGWSPN
Mvan_4302|M.vanbaalenii_PYR-1 VRNFLVANALYWLQEYHIDGLRVDAVASMLY------LDYSRPEGGWSPN
MUL_3936|M.ulcerans_Agy99 VRNFLVANALYWIEQFHIDGLRVDAVASMLY------LDYSRPQDGWTPN
MMAR_4072|M.marinum_M VRNFLVANALYWIEQFHIDGLRVDAVASMLY------LDYSRPQDGWTPN
Mb1361c|M.bovis_AF2122/97 VRNFLVANALYWLQEFHIDGLRVDAVASMLY------LDYSRPEGGWTPN
Rv1326c|M.tuberculosis_H37Rv VRNFLVANALYWLQEFHIDGLRVDAVASMLY------LDYSRPEGGWTPN
MSMEG_4918|M.smegmatis_MC2_155 VRNFLVANALYWLQEFHIDGLRVDAVASMLY------LDYSRPEGGWTPN
TH_1050|M.thermoresistible__bu VRNFLVANALYWLQEFHIDGLRVDAVASMLY------LDYSRPEGGWTPN
MAB_1467c|M.abscessus_ATCC_199 VRNFLVANALFWLDQFHIDGLRVDAVASMLY------LDYSRPQGGWTPN
MAV_3212|M.avium_104 VRRYIIGCALRWMRDFHADGLRLDAVHALVDTTAIHILEELATETDWLAT
**.:::. ** *: ::* ****:*** ::: *: .: .* ..
Mflv_2343|M.gilvum_PYR-GCK IYGGRENLEAVQFLQEMNATVHKINPGIVTIAEESTSWPGVTRPTNLG--
Mvan_4302|M.vanbaalenii_PYR-1 IYGGRENLEAVQFLQEMNATVHKINPGIVTIAEESTSWPGVTRPTNLG--
MUL_3936|M.ulcerans_Agy99 IYEGRENLEAVQFLQEMNATAHKAAPGIVTIAEESTSWPGVTRPTSIG--
MMAR_4072|M.marinum_M IYGGRENLEAVQFLQEMNATAHKAAPGIVTIAEESTSWPGVTRPTSIG--
Mb1361c|M.bovis_AF2122/97 VHGGRENLEAVQFLQEMNATAHKVAPGIVTIAEESTSWPGVTRPTNIG--
Rv1326c|M.tuberculosis_H37Rv VHGGRENLEAVQFLQEMNATAHKVAPGIVTIAEESTPWSGVTRPTNIG--
MSMEG_4918|M.smegmatis_MC2_155 QYGGRENLEAVQFLQEMNATVHKVAPGIVTVAEESTSWPGVTRPTNLG--
TH_1050|M.thermoresistible__bu VHGGRENLEAVQFLQEMNATVHKVAPGIVTIAEESTSWPGVTRPTNLG--
MAB_1467c|M.abscessus_ATCC_199 IHGGRENLEAVQFLQEMNATVHKLHPGIVTVAEESTSWPGVTRATSLG--
MAV_3212|M.avium_104 QLGRPLSLIAESDLNDPRLITARERGGYGLTAQWADDIHHAIHTAVSGER
.* * . *:: . . : * *: : . :.: *
Mflv_2343|M.gilvum_PYR-GCK -----GLGFSMKWNMGWMNDTLEFIKRDPIHRSFHHGEITFSMIYAFSEN
Mvan_4302|M.vanbaalenii_PYR-1 -----GLGFSMKWNMGWMNDTLEFIKRDPIHRSYHHHEMTFSLLYAFSEN
MUL_3936|M.ulcerans_Agy99 -----GLGFSMKWNMGWMHDTLDYVSRDPIYRSYHHHEMTFSMLYAFSEN
MMAR_4072|M.marinum_M -----GLGFSMKWNMGWMHDTLDYVGRDPIYRSYHHHEMTFSMLYAFSEN
Mb1361c|M.bovis_AF2122/97 -----GLGFSMKWNMGWMHDTLDYVSRDPVYRSYHHHEMTFSMLYAFSEN
Rv1326c|M.tuberculosis_H37Rv -----GLGFSMKWNMGWMHDTLDYVSRDPVYRSYHHHEMTFSMLYAFSEN
MSMEG_4918|M.smegmatis_MC2_155 -----GLGFSMKWNMGWMNDTLAYISRDPIYRSYHHHEMTFSMLYAYSEN
TH_1050|M.thermoresistible__bu -----GLGFSMKWNMGWMNDTLEYMRRDPIYRSYHHHEMTFSMLYAFSEN
MAB_1467c|M.abscessus_ATCC_199 -----GLGFSMKWNMGWMHDTLGYLGRDPIHRSFHHHEMTFSMLYAFSEN
MAV_3212|M.avium_104 QGYYADFGSIATLAHTLRHGYFHAATYSSFRRRRHGRPLDTSAQTGIPAT
.:* . :. : ... * * : * . . .
Mflv_2343|M.gilvum_PYR-GCK FVLPISHDEVVHGKGTLWGRMPGNDHMKAAGIRSLLAYQWAHPGKQLLFM
Mvan_4302|M.vanbaalenii_PYR-1 FVLPISHDEVVHGKGTLWGRMPGNDHMKAAGIRSLLAYQWAHPGKQLLFM
MUL_3936|M.ulcerans_Agy99 YVLPLSHDEVVHGKGTLWGRMPGSNHTKAAGLRSLLAYQWAHPGKQLLFM
MMAR_4072|M.marinum_M YVLPLSHDEVVHGKGTLWGRMPGSNHTKAAGLRSLLAYQWAHPGKQLLFM
Mb1361c|M.bovis_AF2122/97 YVLPLSHDEVVHGKGTLWGRMPGNNHVKAAGLRSLLAYQWAHPGKQLLFM
Rv1326c|M.tuberculosis_H37Rv YVLPLSHDEVVHGKGTLWGRMPGNNHVKAAGLRSLLAYQWAHPGKQLLFM
MSMEG_4918|M.smegmatis_MC2_155 YVLPISHDEVVHGKGTLWSRMPGGDHEKAAGLRSLLAYQWAHPGKQLLFM
TH_1050|M.thermoresistible__bu YVLPISHDEVVHGKGTLWTRMPGDDHAKAAGLRSLLAYQWAHPGKQLLFM
MAB_1467c|M.abscessus_ATCC_199 FVLPISHDEVVHGKGTLWTRIPGDGHAKAAGLRCLLAYMWAHPGKQLLFM
MAV_3212|M.avium_104 RLLAYTCTHDQVGNRALGDRPSQNLTAGQLAVKAVLALLSPYT--AMLFM
:*. : . *: :* * . . .::.:** .:. :***
Mflv_2343|M.gilvum_PYR-GCK GQEFGQR---AEWSEERGVDWFQLDEQGFSDGILRMLSDANHIYRDRRAL
Mvan_4302|M.vanbaalenii_PYR-1 GQEFGQR---AEWSEERGVDWFQLDEQGFSGGILRMMTDANEIYRSRRAL
MUL_3936|M.ulcerans_Agy99 GQEFGQR---AEWSEQRGLDWFQLDENGFSNGVLRLVRDINEIYCDHCAL
MMAR_4072|M.marinum_M GQEFGQR---AEWSEQRGLDWFQLDENGFSNGVLRLVRDINEIYCDHCAL
Mb1361c|M.bovis_AF2122/97 GQEFGQR---AEWSEQRGLDWFQLDENGFSNGIQRLVRDINDIYRCHPAL
Rv1326c|M.tuberculosis_H37Rv GQEFGQR---AEWSEQRGLDWFQLDENGFSNGIQRLVRDINDIYRCHPAL
MSMEG_4918|M.smegmatis_MC2_155 GQEFGQR---AEWSEERGLDWYQLDEQSFSTGVQNLVRDLNAIYRSRPAL
TH_1050|M.thermoresistible__bu GQEFGQR---AEWSEERGVDWFQLDEDGFSSGIAQLVRDINRIYQSRRAL
MAB_1467c|M.abscessus_ATCC_199 GQEFGQR---AEWSDERGVDWFQLSEDGYSAGIAALTADLNTVYRERRAL
MAV_3212|M.avium_104 GEEYGASTPFQFFSSHPEPELARATAEGRKTEFAEHGWDASKKGEIPDPQ
*:*:* :*.. : : :. . . * . .
Mflv_2343|M.gilvum_PYR-GCK WSRDTQPEGYSWIDANDSANNVLSFLR------FGDDGSMMACVFNFSGA
Mvan_4302|M.vanbaalenii_PYR-1 WSRDTQPEGYSWIDANDSANNVLSFLR------FGDDGSMMACVFNFSGS
MUL_3936|M.ulcerans_Agy99 WSQDTTPEGYSWIDANDSANNVLSFLR------HGSDGSVMACIFNFAGS
MMAR_4072|M.marinum_M WSQDTTPEGYSWIDANDSANNVLSFLR------HGSDGSVMACIFNFAGS
Mb1361c|M.bovis_AF2122/97 WSLDTTPEGYSWIDANDSANNVLSFMR------YGSDGSVLACVFNFAGA
Rv1326c|M.tuberculosis_H37Rv WSLDTTPEGYSWIDANDSANNVLSFMR------YGSDGSVLACVFNFAGA
MSMEG_4918|M.smegmatis_MC2_155 WSRDTSPEGYSWIDANDSANNVLSFLR------FGNDGSMLACVINFAGS
TH_1050|M.thermoresistible__bu WSRDSSHEGYSWIDANDSANNVLSFLR------FGDDGSMLACVFNFSGT
MAB_1467c|M.abscessus_ATCC_199 WSQDTAPQGYSWIDANDSANNVLSFLR------FGDDGSVLACIFNFAGV
MAV_3212|M.avium_104 DPQTFARSKLNWDEVGTGEHARLHRLYRDLIALRHNDPDLADPWLEHLTV
. . .* :.. . : * : .* .: ::.
Mflv_2343|M.gilvum_PYR-GCK E-HSRYRLG--LPHAGTWREVLNTDSDTYHGAGIGNYGSVEATDEPWHGR
Mvan_4302|M.vanbaalenii_PYR-1 E-HTRYRLG--LPHAGRWREVLNTDSDTYHGTGIGNYGAVEATDEPWHGR
MUL_3936|M.ulcerans_Agy99 E-HSDYRLG--LPIAGRWREVLNTDATIYNGSGVGNFGGVDATAEPWHGR
MMAR_4072|M.marinum_M E-HSDYRLG--LPIAGRWREVLNTDATIYNGSGVGNFGGVDATAEPWHGR
Mb1361c|M.bovis_AF2122/97 E-HRDYRLG--LPRAGRWREVLNTDATIYHGSGIGNLGGVDATDDPWHGR
Rv1326c|M.tuberculosis_H37Rv E-HRDYRLG--LPRAGRWREVLNTDATIYHGSGIGNLGGVDATDDPWHGR
MSMEG_4918|M.smegmatis_MC2_155 E-HSQYRLG--LPHAGTWREVLNTDADIYNGSGIGNYGAVEATDEPWHGR
TH_1050|M.thermoresistible__bu XXXPGVRRGQPLPADRGRRLPLAAIDQIWLRAGLAIFSAIGAATLLIGAA
MAB_1467c|M.abscessus_ATCC_199 E-HSSYRVG--LPLTGRWREVINTDALQYRGGGVGNLGEVIASANPWHGR
MAV_3212|M.avium_104 DYDEDQRWIVLARGRLRIVCNLGAEPVTVPVGGELMLAWDEPTVDADTTA
* : : * . .:
Mflv_2343|M.gilvum_PYR-GCK PASAVMVLPPLSALWFEPVPPEA
Mvan_4302|M.vanbaalenii_PYR-1 PASAVMVLPPLSALWFEPE----
MUL_3936|M.ulcerans_Agy99 PASAVLVLPPSSALWLEPA----
MMAR_4072|M.marinum_M PASAVLVLPPSSALWLEPA----
Mb1361c|M.bovis_AF2122/97 PASAVLVLPPTSALWLTPA----
Rv1326c|M.tuberculosis_H37Rv PASAVLVLPPTSALWLTPA----
MSMEG_4918|M.smegmatis_MC2_155 PASAVMVLPPLSMLWFEPA----
TH_1050|M.thermoresistible__bu TYLMAGGTRPAPGGAAMPSPG--
MAB_1467c|M.abscessus_ATCC_199 PASATLALPPAAAIWLEPVSDRL
MAV_3212|M.avium_104 LQGHSFAILSTPGQPVDN-----
. .