For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GGTFDPIHYGHLVAASEVADRYELDEVVFVPSGQPWQKGRRVSPAEDRYLMTVIATASNPRFSVSRVDID RGGPTYTKDTLRDLHALNPAAELYFITGADALASIMSWQGWEQMFESARFVGVSRPGYELRHDHVTAALD GLAENALSLVEIPALAISSTDCRRRAAHGRPLWYLMPDGVVQYVSKRRLYRTPPDSGGRGDPGHHDHSAR GEQHMSATQEAIDMATVAAGAASAKLADDVLVIDVSGQLVITDCFVIASASNERQVNAIVDEVEEKMRRA GYKLARREGAREGRWTLLDYRDIVVHVQHQDDRSFYALDRLWADCPVVAVDLAEGSDDPAEAP*
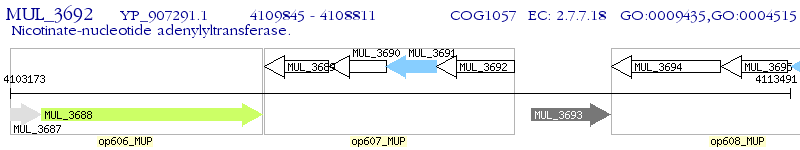
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3692 | nadD | - | 100% (344) | bifunctional nicotinate-nucleotide adenylyltransferase NadD/hypothetical protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2444c | nadD | 4e-93 | 82.00% (200) | nicotinic acid mononucleotide adenylyltransferase |
| M. gilvum PYR-GCK | Mflv_2653 | nadD | 3e-84 | 76.38% (199) | nicotinate (nicotinamide) nucleotide adenylyltransferase |
| M. tuberculosis H37Rv | Rv2421c | nadD | 9e-94 | 82.50% (200) | nicotinic acid mononucleotide adenylyltransferase |
| M. leprae Br4923 | MLBr_01454 | nadD | 1e-90 | 78.11% (201) | nicotinic acid mononucleotide adenylyltransferase |
| M. abscessus ATCC 19977 | MAB_1621 | - | 3e-81 | 72.20% (205) | nicotinate-nucleotide adenylyltransferase |
| M. marinum M | MMAR_3749 | nadD | 1e-114 | 99.50% (199) | nicotinate-nucleotide adenylyltransferase NadD |
| M. avium 104 | MAV_1750 | nadD | 1e-93 | 83.59% (195) | nicotinic acid mononucleotide adenylyltransferase |
| M. smegmatis MC2 155 | MSMEG_4581 | nadD | 3e-84 | 72.99% (211) | nicotinate (nicotinamide) nucleotide adenylyltransferase |
| M. thermoresistible (build 8) | TH_0789 | - | 2e-85 | 76.29% (194) | PROBABLE NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE NADD |
| M. vanbaalenii PYR-1 | Mvan_3926 | nadD | 2e-84 | 76.80% (194) | nicotinate (nicotinamide) nucleotide adenylyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3692|M.ulcerans_Agy99 -------------MGGTFDPIHYGHLVAASEVADRYELDEVVFVPSGQPW
MMAR_3749|M.marinum_M -------------MGGTFDPIHYGHLVAASEVADRYELDEVVFVPSGQPW
MLBr_01454|M.leprae_Br4923 -------------MGGTFDPIHYGHLVAASEVAHMFELDEVMFVPSGQPW
MAV_1750|M.avium_104 ------------MMGGTFDPIHYGHLVAASEVADLFGLDQVVFVPSGQPW
Mb2444c|M.bovis_AF2122/97 -------------MGGTFDPIHYGHLVAASEVADLFDLDEVVFVPSGQPW
Rv2421c|M.tuberculosis_H37Rv -------------MGGTFDPIHYGHLVAASEVADLFDLDEVVFVPSGQPW
Mflv_2653|M.gilvum_PYR-GCK -------------MGGTFDPIHNGHLVAASEVADLFDLHEVVFVPTGQPW
Mvan_3926|M.vanbaalenii_PYR-1 MQAGGRRQRRLGVMGGTFDPIHNGHLVAASEVADRFDLDEVVFVPTGQPW
MSMEG_4581|M.smegmatis_MC2_155 ----MATRRRLGVMGGTFDPIHNGHLVAASEVADLFDLDEVVFVPTGEPW
MAB_1621|M.abscessus_ATCC_1997 ---MQSERRRLGVMGGTFDPIHNGHLVAASEVADRFALDEVIFVPTGQPW
TH_0789|M.thermoresistible__bu -------------MGGTFDPIHHGHLVAASEVADLFDLDEVLFVPTGQPW
********* **********. : *.:*:***:*:**
MUL_3692|M.ulcerans_Agy99 QKG-RRVSPAEDRYLMTVIATASNPRFSVSRVDIDRGGPTYTKDTLRDLH
MMAR_3749|M.marinum_M QKG-RRVSPAEDRYLMTVIATASNPRFSVSRVDIDRGGPTYTKDTLRDLH
MLBr_01454|M.leprae_Br4923 QKG-RHVSAAEDRYLMTVIATASNPRFSVSRVDIDRTGPTYTRDTVHDLH
MAV_1750|M.avium_104 QKD-RHVSAAEDRYLMTVIATASNPRFSVSRVDIDRAGPTYTRDTLRDLH
Mb2444c|M.bovis_AF2122/97 QKG-RQVSAAEHRYLMTVIATASNPRFSVSRVDIDRGGPTYTKDTLADLH
Rv2421c|M.tuberculosis_H37Rv QKG-RQVSAAEHRYLMTVIATASNPRFSVSRVDIDRGGPTYTKDTLADLH
Mflv_2653|M.gilvum_PYR-GCK QKRSRPVTPAEDRYLMTVIATASNPRFSVSRVDIDRGGATYTKDTLRDLR
Mvan_3926|M.vanbaalenii_PYR-1 QKRARAVTAAEDRYLMTVIATASNPRFTVSRVDIDRGGATYTKDTLRDLR
MSMEG_4581|M.smegmatis_MC2_155 QKHHRRVSAAEDRYLMTVIATASNPRFSVSRVDIDRGGPTYTKDTLRDLR
MAB_1621|M.abscessus_ATCC_1997 QKQGRKVSPAEHRYLMTVIATASNPRFTVSRADIDRGGATYTVDTLTDLR
TH_0789|M.thermoresistible__bu QKH-REVTAAEDRYLMTVIATASNPRFGVSRVDIDRPGPTYTRDTLRDLK
** * *:.**.*************** ***.**** *.*** **: **:
MUL_3692|M.ulcerans_Agy99 ALNPAAELYFITGADALASIMSWQGWEQMFESARFVGVSRPGYELRHDHV
MMAR_3749|M.marinum_M ALNPAAELYFITGADALASIMSWQGWEQMFESARFVGVSRPGYELRHDHV
MLBr_01454|M.leprae_Br4923 ALNPDSELYFITGADALASILSWQGWDELFDLARFVGVSRPGYELGQEHI
MAV_1750|M.avium_104 ALNPDSELFFITGADALASILSWQGWETLFELAHFVGVSRPGYELRREHI
Mb2444c|M.bovis_AF2122/97 ALHPDSELYFTTGADALASIMSWQGWEELFELARFVGVSRPGYELRNEHI
Rv2421c|M.tuberculosis_H37Rv ALHPDSELYFTTGADALASIMSWQGWEELFELARFVGVSRPGYELRNEHI
Mflv_2653|M.gilvum_PYR-GCK AQNPDADLYFITGADALASILSWQNWEEMFAIARFIGVSRPGYELDGKHI
Mvan_3926|M.vanbaalenii_PYR-1 AQNPDADLFFITGADALASILSWQNWEEMFSIARFIGVSRPGYELDGKHI
MSMEG_4581|M.smegmatis_MC2_155 DLNTDADLYFITGADALGSILSWQNWEDMFSMAKFVGVSRPGYELDGKHI
MAB_1621|M.abscessus_ATCC_1997 TAHPDADLYFITGADALASILSWENWEQLFTLAKFIGVSRPGYELSSDHI
TH_0789|M.thermoresistible__bu TMHPDAELYFITGADALASILSWQDWEEMFSLARFVGVSRPGYELDGEHI
:. ::*:* ******.**:**:.*: :* *:*:********* .*:
MUL_3692|M.ulcerans_Agy99 TAALDGLAENALSLVEIPALAISSTDCRRRAAHGRPLWYLMPDGVVQYVS
MMAR_3749|M.marinum_M TAALDGLAEDALSLVEIPALAISSTDCRRRAAHGRPLWYLMPDGVVQYVS
MLBr_01454|M.leprae_Br4923 TGVMGELPADALTLVEIPALAISSTDCRQRVEQRQPLWYLMPDGVVQYVF
MAV_1750|M.avium_104 TGVLGELPDDALTLVEIPALAISSTDCRQRAAHRRPLWYLMPDGVVQYVS
Mb2444c|M.bovis_AF2122/97 TSLLGQLAKDALTLVEIPALAISSTDCRQRAEQSRPLWYLMPDSVVQYVS
Rv2421c|M.tuberculosis_H37Rv TSLLGQLAKDALTLVEIPALAISSTDCRQRAEQSRPLWYLMPDGVVQYVS
Mflv_2653|M.gilvum_PYR-GCK SAAMAELPADALHLVEVPALAISSTDCRLRAEQSRPIWYLVPDGVVQYVA
Mvan_3926|M.vanbaalenii_PYR-1 SAAMAELPDDALHLIEVPALAISSTDCRIRAEQSRPIWYLVPDGVVQYVA
MSMEG_4581|M.smegmatis_MC2_155 LDAMRELPPDALSLVEVPALAISSSDCRKRAEEQRPIWYLVPDGVVQYVA
MAB_1621|M.abscessus_ATCC_1997 --AHAELPPDGLSLVEVPALAISSTDCRIRAGQARPIWYLVPDGVVQYVA
TH_0789|M.thermoresistible__bu LGALKEKPADALTLVEVPALAISSSDCRARAAAGRPIWYLVPDGVVQYVA
. :.* *:*:*******:*** *. :*:***:**.*****
MUL_3692|M.ulcerans_Agy99 KRRLYRTPPDSGGRGDPGHHDHSARGEQHMSATQEAIDMATVAAGAASAK
MMAR_3749|M.marinum_M KRRLYRTPPDSGGG--------------------ETLDTTTILPAGNNT-
MLBr_01454|M.leprae_Br4923 KRRLYRGPHGVAGTG---------------------RLTTPSLVVGNNT-
MAV_1750|M.avium_104 KRRLYRAEPGPAVA-----------------------VTETSLSTGDHP-
Mb2444c|M.bovis_AF2122/97 KCRLYCGACDAGAR------------------------STTSLAAGNGL-
Rv2421c|M.tuberculosis_H37Rv KCRLYCGACDAGAR------------------------STTSLAAGNGL-
Mflv_2653|M.gilvum_PYR-GCK KRDLYRNQN---PA------------------------GEGERP------
Mvan_3926|M.vanbaalenii_PYR-1 KRNLYRSE------------------------------GEGVRP------
MSMEG_4581|M.smegmatis_MC2_155 KRGLYTRKP---NN------------------------GEAKDGDVKDEE
MAB_1621|M.abscessus_ATCC_1997 KHRLYSGNK---GN------------------------QGGLA-------
TH_0789|M.thermoresistible__bu KRKLYQNPDRVVPQ------------------------PSAKRACAKSQG
* **
MUL_3692|M.ulcerans_Agy99 LADDVLVIDVSGQLVITDCFVIASASNERQVNAIVDEVEEKMRRAGYKLA
MMAR_3749|M.marinum_M --------------------------------------------------
MLBr_01454|M.leprae_Br4923 --------------------------------------------------
MAV_1750|M.avium_104 --------------------------------------------------
Mb2444c|M.bovis_AF2122/97 --------------------------------------------------
Rv2421c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_2653|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3926|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4581|M.smegmatis_MC2_155 AVR-----------------------------------------------
MAB_1621|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0789|M.thermoresistible__bu DQ------------------------------------------------
MUL_3692|M.ulcerans_Agy99 RREGAREGRWTLLDYRDIVVHVQHQDDRSFYALDRLWADCPVVAVDLAEG
MMAR_3749|M.marinum_M --------------------------------------------------
MLBr_01454|M.leprae_Br4923 --------------------------------------------------
MAV_1750|M.avium_104 --------------------------------------------------
Mb2444c|M.bovis_AF2122/97 --------------------------------------------------
Rv2421c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_2653|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3926|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4581|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1621|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0789|M.thermoresistible__bu --------------------------------------------------
MUL_3692|M.ulcerans_Agy99 SDDPAEAP
MMAR_3749|M.marinum_M --------
MLBr_01454|M.leprae_Br4923 --------
MAV_1750|M.avium_104 --------
Mb2444c|M.bovis_AF2122/97 --------
Rv2421c|M.tuberculosis_H37Rv --------
Mflv_2653|M.gilvum_PYR-GCK --------
Mvan_3926|M.vanbaalenii_PYR-1 --------
MSMEG_4581|M.smegmatis_MC2_155 --------
MAB_1621|M.abscessus_ATCC_1997 --------
TH_0789|M.thermoresistible__bu --------