For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTHYTWTVIGAGPAGIASVGRLLDHGVRPNEIAWIDPQFAAGDLGEKWGAVPSNTQVSTFLDYFNASPAF GFAAAPHFDLHDIDPQHTCQLGVVAGPLRWVTEHLRAKVAAFQSTATELSLRNQHWQITTLDGEISSKNV ILAVGSTPKKLTYPELKEIPVEVALNPEKLAQQQLDGATVAVFGSSHSTMIALPNLLEHPVHKVINFYRS PHRYAVPFQDWTLFDDTGLKGDAARWARENIDGWHPERLHRCLVDSPEFARLLRECDQAVYTVGFERRHL PLTPQWGPLEHDGANGILAPGLFGIGIAFPEYRTDPLGFGEHRVGMAKFMQRLNSVLPLWLQYGGSPAQA AALCPDGVGLLAGATA
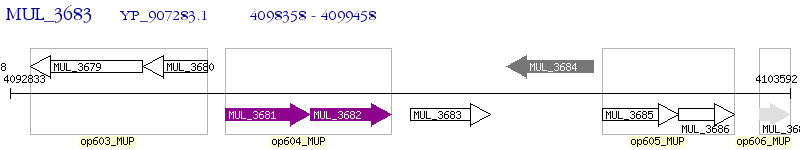
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3683 | - | - | 100% (366) | hypothetical protein MUL_3683 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0670 | - | 1e-136 | 66.86% (344) | hypothetical protein MAB_0670 |
| M. marinum M | MMAR_3739 | - | 0.0 | 98.91% (366) | hypothetical protein MMAR_3739 |
| M. avium 104 | MAV_1699 | - | 1e-127 | 66.17% (337) | hypothetical protein MAV_1699 |
| M. smegmatis MC2 155 | MSMEG_0543 | - | 1e-123 | 59.36% (342) | hypothetical protein MSMEG_0543 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3683|M.ulcerans_Agy99 --MTHYTWTVIGAGPAGIASVGRLLDHGVRPNEIAWIDPQFAAGDLGEKW
MMAR_3739|M.marinum_M --MTHYTWTVIGAGPAGIASVGRLLDHGVRPNEIAWIDPQFAAGDLGEKW
MAV_1699|M.avium_104 ---------MVGAGPAGIAAVGRLLDHGADS--IAWIDPAFAAGDIGQKW
MAB_0670|M.abscessus_ATCC_1997 --MSEYVWTVVGAGPAGIAAVGKLIDHGVLPASIAWIDPEFTAGDLGAKW
MSMEG_0543|M.smegmatis_MC2_155 MSSAHLRWTVIGAGPAGIATVGRLIDNGIRHDEIAWVDPGFAVGDFGTKW
::********:**:*:*:* ***:** *:.**:* **
MUL_3683|M.ulcerans_Agy99 GAVPSNTQVSTFLDYFNASPAFGFAAAPHFDLHDIDPQHTCQLGVVAGPL
MMAR_3739|M.marinum_M GAVPSNTQVSLFLDYFNASPAFGFAAAPHFDLHDIDPQHTCQLGVVAGPL
MAV_1699|M.avium_104 RSVSSNTHAGLFLEYFNGCKSFRFSEAPPMPLREIDAGETCALALVAEPL
MAB_0670|M.abscessus_ATCC_1997 RAVPSNTSVKLFINYLTGADSFRFAHAPHFALNDLAPTDTCLLGDVADPL
MSMEG_0543|M.smegmatis_MC2_155 SVVPSNTRVSGFVHFLQASAAFQFHRGPDFAIRELDPRQTCSLAMVTEPL
*.*** . *:.:: .. :* * .* : :.:: . .** *. *: **
MUL_3683|M.ulcerans_Agy99 RWVTEHLRAKVAAFQSTATELSLRNQHWQITTLDGEISSKNVILAVGSTP
MMAR_3739|M.marinum_M RWVTEHLRAKVAAFQSTATELSLRNQHWQITTLDGEISSKNVILAVGSTP
MAV_1699|M.avium_104 LWVTGQLRERVDTVTTTATALYLSDRRWRIETEQREIFSRNVILAVGAVP
MAB_0670|M.abscessus_ATCC_1997 VWITGQLCAQVSALRTRATGLALHGGRWEVQTELGEITSKNVILAVGSVP
MSMEG_0543|M.smegmatis_MC2_155 QWITDQLAERVTAVKALATRLELHGRHWEVRTDIATLVSDNVVLAIGSEP
*:* :* :* :. : ** * * . :*.: * : * **:**:*: *
MUL_3683|M.ulcerans_Agy99 KKLTYPELKEIPVEVALNPEKLAQQQLDGATVAVFGSSHSTMIALPNLLE
MMAR_3739|M.marinum_M KKLTYPELKEIPVEVALNPEKLAQQQLDGATVAVFGSSHSTMIALPNLLE
MAV_1699|M.avium_104 KKLCHPGLEEIPVEVALDPEKLARQPLSGATVGVFGSSHSSMIALPNLLR
MAB_0670|M.abscessus_ATCC_1997 KKLAYPWLNEIPIEVALDPAKLAEQPLEGATVAVFGASHSSMIALPNLLA
MSMEG_0543|M.smegmatis_MC2_155 RRLDHPGIQEIPLDVAVAPHKLATESLDGATVAVFGSSHSTMLVLPNLLR
::* :* ::***::**: * *** : *.****.***:***:*:.*****
MUL_3683|M.ulcerans_Agy99 HPVHKVINFYRSPHRYAVPFQDWTLFDDTGLKGDAARWARENIDGWHPER
MMAR_3739|M.marinum_M HPVHKVINFYRSPHRYAVPFQDWTLFDDTGLKGDAARWARENIDGRHPER
MAV_1699|M.avium_104 QPVEKVINFYRSPLKYAVYFDDWILFDDTGLKGQAAVWARENIDGVLPER
MAB_0670|M.abscessus_ATCC_1997 GPAAKVINFYRGPLRYAVDMGEWTLFDDTGLKGEAARWARENIDGVLPDR
MSMEG_0543|M.smegmatis_MC2_155 RPVARVVNFYQHPLRYAVDFGDWTLFDDTGLKGHAAQWARENIDGTLPER
*. :*:***: * :*** : :* *********.** ******** *:*
MUL_3683|M.ulcerans_Agy99 LHRCLVDSPEFARLLRECDQAVYTVGFERRHLPLTPQWGPLEHDGANGIL
MMAR_3739|M.marinum_M LHRCLVDSPEYARLLRECDQAVYTVGFERRHLPLTPQWGPLEHDGANGIL
MAV_1699|M.avium_104 LHRCLVSSPEFAENLARCDRVVYTVGFERRTLPETPQWGRLDYNPTTGIL
MAB_0670|M.abscessus_ATCC_1997 LQRCLVDSPEFPELLESCDYAVYTVGFSPRPIPAAPQWGQLECNAANGII
MSMEG_0543|M.smegmatis_MC2_155 LRRCLVSDPRFDELLAQCDRVVYTVGFQPRRI-DAPQLGSLLHNPANGII
*:****..*.: . * ** .******. * : :** * * : :.**:
MUL_3683|M.ulcerans_Agy99 APGLFGIGIAFPEYRTDPLGFGEHRVGMAKFMQRLNSVLPLWLQYGGSPA
MMAR_3739|M.marinum_M APGLFGIGIAFPEYRTDPLGFGEHRVGMAKFMQRLNSVLPLWLQYGGSPT
MAV_1699|M.avium_104 APGLFGVGIAFPQYAEDPYGYGQFRVGLKKFMDYLDSVLPLWLRYG--P-
MAB_0670|M.abscessus_ATCC_1997 APGLFGVGIAFPEYRIDPTGFGEYRVGLQKFMDRLTKTLPLWLKYGS---
MSMEG_0543|M.smegmatis_MC2_155 APGLFGVGIAFPGFRTDPTGLGEHRVGLQKFMEQLATCMPIWLRYGA---
******:***** : ** * *:.***: ***: * . :*:**:**
MUL_3683|M.ulcerans_Agy99 QAAALCPDGVGLLAGATA
MMAR_3739|M.marinum_M QAAALCPDGVGLLAGATA
MAV_1699|M.avium_104 ------------------
MAB_0670|M.abscessus_ATCC_1997 ------------------
MSMEG_0543|M.smegmatis_MC2_155 ------------------