For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSLQDRLASILRDVLPVEEEPDGALTVRHDGTFASLRVVNIAEDLDLVSLTQILAWDLPLTKKISDLLAN QARDTNFGAVTTVEKVADKAVPRNAGKGSAKTLPKTVDVMLRYNFPGAGLTDNALRTLILLVLDSGGQIR GKLTS*
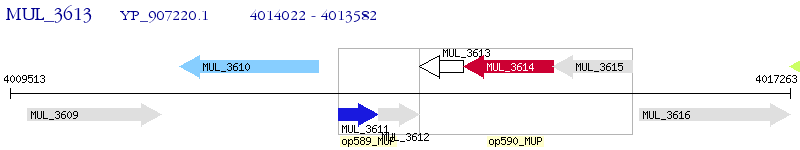
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3613 | - | - | 100% (146) | hypothetical protein MUL_3613 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2381c | - | 2e-53 | 72.22% (144) | hypothetical protein Mb2381c |
| M. gilvum PYR-GCK | Mflv_2715 | - | 9e-38 | 58.16% (141) | hypothetical protein Mflv_2715 |
| M. tuberculosis H37Rv | Rv2360c | - | 2e-53 | 72.22% (144) | hypothetical protein Rv2360c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1677 | - | 1e-20 | 38.57% (140) | hypothetical protein MAB_1677 |
| M. marinum M | MMAR_3670 | - | 2e-76 | 98.63% (146) | hypothetical protein MMAR_3670 |
| M. avium 104 | MAV_2035 | - | 1e-53 | 74.13% (143) | hypothetical protein MAV_2035 |
| M. smegmatis MC2 155 | MSMEG_4489 | - | 3e-36 | 54.55% (154) | hypothetical protein MSMEG_4489 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3821 | - | 8e-41 | 60.99% (141) | hypothetical protein Mvan_3821 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3613|M.ulcerans_Agy99 -------MPSLQDRLASILRDVLPVEEEPDGALTVRHDGTFASLRVVNIA
MMAR_3670|M.marinum_M -------MPSLQDRLASILRDVLPVEEEPDGALTVRHDGTFASLRVVNIA
MAV_2035|M.avium_104 -------MSSLQERLASVLREVLPSQEESDGALTVHHQGTIASLRVVNIA
Mb2381c|M.bovis_AF2122/97 -------MPSLPDRLASILRDVLPAEEEPDGALTVRHDGTFASLRVVSIA
Rv2360c|M.tuberculosis_H37Rv -------MPSLPDRLASILRDVLPAEEEPDGALTVRHDGTFASLRVVSIA
Mflv_2715|M.gilvum_PYR-GCK --------MDLTERLAVVLAEILPVTREDDGALTVQHEQTFASLRVVTIA
Mvan_3821|M.vanbaalenii_PYR-1 --------MELSERLAAVLAEILPVTHEEDGALTVRHDETFASLRVVTIA
MSMEG_4489|M.smegmatis_MC2_155 ----MNAMPDLLQRLSTVLAEVMSVAEEADGALTITSGGSVALVRVVAIA
MAB_1677|M.abscessus_ATCC_1997 MTDPEGAAGKILDRVYRVLHPVLPVTREADGALTADYEGTLTSIRAVTIA
.: :*: :* ::. .* ***** :.: :*.* **
MUL_3613|M.ulcerans_Agy99 EDLDLVSLTQILAWDLPLTKKISDLLANQARDTNFGAVTTVEKVADKAVP
MMAR_3670|M.marinum_M EDLDLVSLTQILAWDLPLTKKIGDLLANQARDTNFGAVTTVEKVADKAVQ
MAV_2035|M.avium_104 EDLDLVSLTQILAWDLPLTKKVSDHVARQARDANFGSVSVVEKVNKTAVQ
Mb2381c|M.bovis_AF2122/97 EDLELVSLTQILAWDLPLTKRLTEQVAKQARDINFGSVSLREKVSEKAAR
Rv2360c|M.tuberculosis_H37Rv EDLELVSLTQILAWDLPLTKRLAEQVAKQARDINFGSVSLREKVSEKAAR
Mflv_2715|M.gilvum_PYR-GCK EGLDLVSLTQVLAWDLPLDSRIRAQVAKHAHDTLLGTVSLVEKVGTTHKG
Mvan_3821|M.vanbaalenii_PYR-1 EDLDLVSLTQVLAWDLPLDSKIRAKVAKHAHDTLLGTVSLVEKIG-AAKG
MSMEG_4489|M.smegmatis_MC2_155 EDLEMVSLTQPLAWDLPLNNKIRERVVGHANRTMLGSVVLVEKVADGPAA
MAB_1677|M.abscessus_ATCC_1997 TGLDVVSLSQVLAWDVAVNAKSRHLVDGLAQKSLFGNILLIAKGR-----
.*::***:* ****:.: : : *. :* : *
MUL_3613|M.ulcerans_Agy99 RNAGKGSAKTL-----PKTVDVMLRYNFPGAGLTDN-ALRTLILLVLDSG
MMAR_3670|M.marinum_M RNAGKGSAKTL-----PKTVDVMLRYNFPGAGLTDN-ALRTLILLVLDSG
MAV_2035|M.avium_104 RNSGKNTAK---------LADVMLRYNFPGAGLTDD-ALRTLVLLVLDTG
Mb2381c|M.bovis_AF2122/97 RSSGRPASN---------TADVMLRYNFPGTGLTDD-ALRTLILLVLETG
Rv2360c|M.tuberculosis_H37Rv RSSGRPASN---------TADVMLRYNFPGTGLTDD-ALRTLILLVLETG
Mflv_2715|M.gilvum_PYR-GCK TAKSKKTGD------------VLLRYNFPAAGLTDD-ALRTLVLMVLATG
Mvan_3821|M.vanbaalenii_PYR-1 --NSKKTGD------------VLLRYNFPAAGLTDD-ALRTLVMMVLATG
MSMEG_4489|M.smegmatis_MC2_155 PVNGTGPKSATRKGSTKKVADVMLRYNFPAAGLTDD-ALRTLILMVLETG
MAB_1677|M.abscessus_ATCC_1997 ------------------KADVLLRYNFPATGIDDDDALVTLLMMVFATG
*:******.:*: *: ** **:::*: :*
MUL_3613|M.ulcerans_Agy99 GQIRGKLTS
MMAR_3670|M.marinum_M GQIRGKLTS
MAV_2035|M.avium_104 ARIRHTLTD
Mb2381c|M.bovis_AF2122/97 ATIRSALVG
Rv2360c|M.tuberculosis_H37Rv ATIRSALVG
Mflv_2715|M.gilvum_PYR-GCK TDVRNAMVG
Mvan_3821|M.vanbaalenii_PYR-1 GDVRRALIA
MSMEG_4489|M.smegmatis_MC2_155 ADVRRDLTS
MAB_1677|M.abscessus_ATCC_1997 ADARKALGN
* :