For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGVWTEGTAADNQASLAELAALAETTGSQVLEGLIQRRDRPDPSTYIGSGKAAELREVVVATGADTVICD GELSPAQLTALEKAVKVKVIDRTALILDIFAQHATSREGKAQVALAQMEYMLPRLRGWGESMSRQAGGRA GGSGGGVGLRGPGETKIETDRRRIRERMAKLRREIKAMKQARDTQRSRRLHSDVPSIAIVGYTNAGKSSL LNALTGAGVLVQDALFATLDPTTRRAEFDPDSNGARPFLFTDTVGFVRHLPPQLVEAFRSTLEEVIDADL LLHVVDGSDPNPLAQINAVRQVISEVIADEATGGGEASDGRAGSAGAEVPQELLVVNKVDAAGDLALAEL RHALPGALFVSAHTGDGIDVLWRKLAELAAPTDTAVDVVIPYDRGDLVARMHADGRVQQAEHQPEGTRIR ARVPGALAATLREFAASSAGPGE*
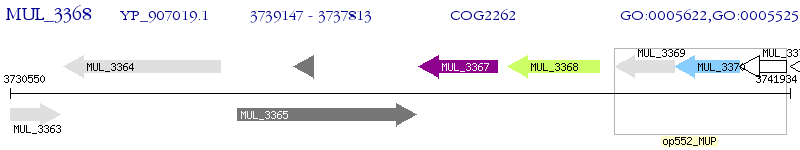
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3368 | hflX | - | 100% (444) | GTP-binding protein HflX |
| M. ulcerans Agy99 | MUL_3232 | engA | 8e-05 | 30.63% (111) | GTP-binding protein EngA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2744c | hflX | 0.0 | 84.63% (436) | GTP-binding protein HflX |
| M. gilvum PYR-GCK | Mflv_3960 | - | 0.0 | 76.43% (437) | GTP-binding protein, HSR1-related |
| M. tuberculosis H37Rv | Rv2725c | hflX | 0.0 | 84.63% (436) | GTP-binding protein HflX |
| M. leprae Br4923 | MLBr_00997 | - | 0.0 | 84.21% (437) | putative ATP/GTP-binding protein |
| M. abscessus ATCC 19977 | MAB_3042c | - | 1e-180 | 76.39% (432) | GTP-binding protein HflX |
| M. marinum M | MMAR_1988 | hflX | 0.0 | 99.10% (444) | GTP-binding protein HflX |
| M. avium 104 | MAV_3618 | - | 0.0 | 83.98% (437) | GTPase |
| M. smegmatis MC2 155 | MSMEG_2736 | - | 0.0 | 77.06% (436) | GTP-binding protein |
| M. thermoresistible (build 8) | TH_1520 | hflX | 0.0 | 77.06% (436) | PROBABLE GTP-BINDING PROTEIN HFLX |
| M. vanbaalenii PYR-1 | Mvan_2436 | - | 0.0 | 77.29% (436) | GTP-binding protein, HSR1-related |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3368|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1988|M.marinum_M ----------MTYSEFPDS-----VRRASQP---AAEPSIGELALEDRSA
MLBr_00997|M.leprae_Br4923 ----------MKYPDSSDRGSADFPDLWDSS---DSVPSTGELALHDRLA
Mb2744c|M.bovis_AF2122/97 MPANSDARPAATCHHRVLAMTYPDPPQTGLS---DFTPSLGELALEDRSA
Rv2725c|M.tuberculosis_H37Rv MPANSDARPAATCHHRVLAMTYPDPPQTGLS---DFTPSLGELALEDRSA
MAV_3618|M.avium_104 ------------MHHLLLPMTHPEFRDDAPA---ITEPSTGELALEDRSA
Mflv_3960|M.gilvum_PYR-GCK -------MRSDIRANLDSPMTYPEFPH--------ETPSVGELALEDRTA
Mvan_2436|M.vanbaalenii_PYR-1 -------MLSDIRANLDSPMTYPEFSN--------DNPSVGELALEDRTA
MSMEG_2736|M.smegmatis_MC2_155 -------------------MTYPENS---------VAPSTGELALEDRAS
TH_1520|M.thermoresistible__bu -------------------MTYPETPETPETVRTAEAPSVGELELQDRAA
MAB_3042c|M.abscessus_ATCC_199 -----------------MRTTY-------------ETPTDGELALEDRAA
MUL_3368|M.ulcerans_Agy99 ----------------------------MVGVWTEGTAADNQASLAELAA
MMAR_1988|M.marinum_M LRRVAGLSTELADVSEVEYRQLRLERVVLVGVWTEGTAADNQASLAELAA
MLBr_00997|M.leprae_Br4923 LRRIAGLSTELADVSEVEYRQLRLERVVLVGVWTEGSAADNQASMAELAT
Mb2744c|M.bovis_AF2122/97 LRRVAGLSTELADVSEVEYRQLRLERVVLVGVWTEGSAADNRASLAELAA
Rv2725c|M.tuberculosis_H37Rv LRRVAGLSTELADVSEVEYRQLRLERVVLVGVWTEGSAADNRASLAELAA
MAV_3618|M.avium_104 LRRVAGLSTELTDISEVEYRQLRLERVVLVGVWTDGTAADSRASMAELAA
Mflv_3960|M.gilvum_PYR-GCK LRRVAGLSTELADISEVEYRKLRLERVVLVGVWTDGSAADADASLAELAA
Mvan_2436|M.vanbaalenii_PYR-1 LRRVAGLSTELTDVSEVEYRQLRLERVVLVGVWTEGSAADADASLAELAA
MSMEG_2736|M.smegmatis_MC2_155 LRRVAGLSTELADVSEVEYRQLRLERVVLVGVWTEGSAADAEASLAELAA
TH_1520|M.thermoresistible__bu LRRVAGLSTELTDVSEVEYRQLRLERVVLVGVWTEGTIADAEASLAELAR
MAB_3042c|M.abscessus_ATCC_199 LKRVAGLSTELADVTEVEYRQLRLERVVLVGVWTEGTSQEAEASMAELAA
:*****:*: : **:****
MUL_3368|M.ulcerans_Agy99 LAETTGSQVLEGLIQRRDRPDPSTYIGSGKAAELREVVVATGADTVICDG
MMAR_1988|M.marinum_M LAETAGSQVLEGLIQRRDRPDPSTYIGSGKAAELREVVVATGADTVICDG
MLBr_00997|M.leprae_Br4923 LAETAGSQVLEGLIQRRDRPDPSTYIGSGKAAELREVVLATGADTVICDG
Mb2744c|M.bovis_AF2122/97 LAETAGSQVLEGLIQRRDKPDPSTYIGSGKAAELREVIVATGADTVICDG
Rv2725c|M.tuberculosis_H37Rv LAETAGSQVLEGLIQRRDKPDPSTYIGSGKAAELREVIVATGADTVICDG
MAV_3618|M.avium_104 LAETAGSQVLEGLIQRRDKPDPSTYIGSGKAAELRDVVLATGADTVICDG
Mflv_3960|M.gilvum_PYR-GCK LAETAGSEVLEGLIQRRDKPDPSTYIGSGKALELRDIVVATGADTVICDG
Mvan_2436|M.vanbaalenii_PYR-1 LAETAGSEVLDGLIQRRDKPDPSTYIGSGKALELREIVVATGADTVICDG
MSMEG_2736|M.smegmatis_MC2_155 LAETAGSEVLEGLIQRRDKPDPSTYIGSGKAAELREVVLATGADTVICDG
TH_1520|M.thermoresistible__bu LAETAGSQVLEGVIQRRDKPDPATYIGSGKARELRDIVVATGADTVICDG
MAB_3042c|M.abscessus_ATCC_199 LAETAGSEVLEGLIQRRQKPDPATYIGSGKAIELREIVLATGADTVICDG
****:**:**:*:****::***:******** ***::::***********
MUL_3368|M.ulcerans_Agy99 ELSPAQLTALEKAVKVKVIDRTALILDIFAQHATSREGKAQVALAQMEYM
MMAR_1988|M.marinum_M ELSPAQLTALEKAVKVKVIDRTALILDIFAQHATSREGKAQVALAQMEYM
MLBr_00997|M.leprae_Br4923 ELSPAQLTALEKAVKVKVIDRTALILDIFAQHATSREGKAQVLLAQMAYM
Mb2744c|M.bovis_AF2122/97 ELSPAQLTALEKAVQVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
Rv2725c|M.tuberculosis_H37Rv ELSPAQLTALEKAVQVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
MAV_3618|M.avium_104 ELSPAQLTALEKAVKVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
Mflv_3960|M.gilvum_PYR-GCK ELSPAQLNALEKVVKVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
Mvan_2436|M.vanbaalenii_PYR-1 ELSPAQLNALEKVVKVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
MSMEG_2736|M.smegmatis_MC2_155 ELSPAQLNALEKAVKVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
TH_1520|M.thermoresistible__bu ELSPAQLTALEKVVQVKVVDRTALILDIFAQHATSREGKAQVSLAQMEYM
MAB_3042c|M.abscessus_ATCC_199 ELSPAQLVALEKAVKVKVIDRTALILDIFAQHATSREGKAQVSLAQMEYM
******* ****.*:***:*********************** **** **
MUL_3368|M.ulcerans_Agy99 LPRLRGWGESMSRQAGGRAGGSGGGVGLRGPGETKIETDRRRIRERMAKL
MMAR_1988|M.marinum_M LPRLRGWGESMSRQAGGRAGGSGGGVGLRGPGETKIETDRRRIRERMAKL
MLBr_00997|M.leprae_Br4923 LPRLRGWGESMSRQVGGRAGGSGGGVGLRGPGETKIETDRRRIRERMAKL
Mb2744c|M.bovis_AF2122/97 LPRLRGWGESMSRQAGGRAGGSGGGVGLRGPGETKIETDRRRIRERMAKL
Rv2725c|M.tuberculosis_H37Rv LPRLRGWGESMSRQAGGRAGGSGGGVGLRGPGETKIETDRRRIRERMAKL
MAV_3618|M.avium_104 LPRLRGWGESMSRQAGGRAGGSGGGVGLRGPGETKIETDRRRIRERMAKL
Mflv_3960|M.gilvum_PYR-GCK LPRLRGWGESMSRQAGGRAGGAGGGVGTRGPGETKIETDRRRIRERMSKL
Mvan_2436|M.vanbaalenii_PYR-1 LPRLRGWGESMSRQAGGRAGGAGGGVGTRGPGETKIETDRRRIRERMAKL
MSMEG_2736|M.smegmatis_MC2_155 LPRLRGWGESMSRQAGGRAGGAGGGVGTRGPGETKIETDRRRIRERMAKL
TH_1520|M.thermoresistible__bu LPRLRGWGESMSRQAGGRAGGSGGGVGLRGPGETKIETDRRRIRERMARL
MAB_3042c|M.abscessus_ATCC_199 LPRLRGWGESMSRQAGGRAGGAGGGVGTRGPGETKIETDRRRIRERMSKL
**************.******:***** *******************::*
MUL_3368|M.ulcerans_Agy99 RREIKAMKQARDTQRSRRLHSDVPSIAIVGYTNAGKSSLLNALTGAGVLV
MMAR_1988|M.marinum_M RREIKAMKQARDTQRSRRLHSDVPSIAIVGYTNAGKSSLLNALTGAGVLV
MLBr_00997|M.leprae_Br4923 RRDIGAMKQARDTQRSRRLHSDIPSIAIVGYTNAGKSSVLNALTGAWVLV
Mb2744c|M.bovis_AF2122/97 RRDIRAMKQVRDTQRSRRRHSDVPSIAIVGYTNAGKSSLLNALTGAGVLV
Rv2725c|M.tuberculosis_H37Rv RRDIRAMKQVRDTQRSRRRHSDVPSIAIVGYTNAGKSSLLNALTGAGVLV
MAV_3618|M.avium_104 RREIKDMKQVRDTQRSRRLHSDVPSIAIVGYTNAGKSSLLNALTGAGVLV
Mflv_3960|M.gilvum_PYR-GCK RREIRDMKQVRDTQRSRRLANDVPSVTIVGYTNAGKSSLLNALTGAGVLV
Mvan_2436|M.vanbaalenii_PYR-1 RREIKDMKKIRDTQRSGRRRTEIPSIAIVGYTNAGKSSLLNALTGAGVLV
MSMEG_2736|M.smegmatis_MC2_155 RREIRDMKKIRDTQRGSRRRSEIPSVAIVGYTNAGKSSLLNALTGAGVLV
TH_1520|M.thermoresistible__bu RREIKEMKKVRDTKRHRRLASAIPSIAIVGYTNAGKSSLLNALTDAGVLV
MAB_3042c|M.abscessus_ATCC_199 RREIRDMKKVRDTKRSRRLESDVPSVAIVGYTNAGKSSLLNAITGAGVLV
**:* **: ***:* * . :**::***********:***:*.* ***
MUL_3368|M.ulcerans_Agy99 QDALFATLDPTTRRAEFDPDSNGARPFLFTDTVGFVRHLPPQLVEAFRST
MMAR_1988|M.marinum_M QDALFATLDPTTRRAEFDPDSNGARPFLFTDTVGFVRHLPTQLVEAFRST
MLBr_00997|M.leprae_Br4923 QDALFVTLEPTTRHAEFD---NGQ-PFVFTDTVGFVRHLPTQLVEAFRST
Mb2744c|M.bovis_AF2122/97 QDALFATLEPTTRRAEFG---DGR-PVVLTDTVGFVRHLPTQLVEAFRST
Rv2725c|M.tuberculosis_H37Rv QDALFATLEPTTRRAEFG---DGR-PVVLTDTVGFVRHLPTQLVEAFRST
MAV_3618|M.avium_104 QDALFATLEPTTRRAEWD---DGR-AFVLTDTVGFVRHLPTQLVEAFRST
Mflv_3960|M.gilvum_PYR-GCK ENALFATLEPTTRRGELS---DGR-EFVLTDTVGFVRHLPTQLVEAFRST
Mvan_2436|M.vanbaalenii_PYR-1 ENALFATLEPTTRRGQFD---DGR-PFVLTDTVGFVRHLPTQLVEAFRST
MSMEG_2736|M.smegmatis_MC2_155 ENALFATLEPTTRRGEFE---DGR-PFVLTDTVGFVRHLPTQLVEAFRST
TH_1520|M.thermoresistible__bu EDALFATLEPTTRRGRFD---DGR-PFVLTDTVGFVRHLPTQLVEAFRST
MAB_3042c|M.abscessus_ATCC_199 QDALFATLEPTTRRGTFD---DGR-EFVITDTVGFVRHLPTQLVEAFRST
::***.**:****:. :* .::***********.*********
MUL_3368|M.ulcerans_Agy99 LEEVIDADLLLHVVDGSDPNPLAQINAVRQVISEVIADEATGGGEASDGR
MMAR_1988|M.marinum_M LEEVIDADLLLHVVDGSDPNPLAQINAVRQVISEVIADEATGGGEASDGR
MLBr_00997|M.leprae_Br4923 LEEVVDADLLLHVVDGSDANPLAQINAVRQVIFEVISDHQDGG-------
Mb2744c|M.bovis_AF2122/97 LEEVVHADLLVHVVDGSDGHPLAQIDAVRQVISEVIADHDGDP-------
Rv2725c|M.tuberculosis_H37Rv LEEVVHADLLVHVVDGSDGHPLAQIDAVRQVISEVIADHDGDP-------
MAV_3618|M.avium_104 LEEVVDADLLVHVVDGSDVNPLAQIDAVHQVISEVIADHHGEP-------
Mflv_3960|M.gilvum_PYR-GCK LEEVVDAELLLHVVDGSDANPLAQINAVRTVVNEVVAETDATA-------
Mvan_2436|M.vanbaalenii_PYR-1 LEEVVDAELLVHVVDGSDVNPLAQINAVRQVINEVAAEYDIAP-------
MSMEG_2736|M.smegmatis_MC2_155 LEEVVDADLLIHVVDGSDVNPLAQINAVRTVINEVVAEYDIAP-------
TH_1520|M.thermoresistible__bu LEEVIHADLLVHVVDGADANPLAQIDAVREVLRDVHADHDAPP-------
MAB_3042c|M.abscessus_ATCC_199 LEEVADADLLVHVVDGSDMAPLAQIEAVRTVIGEVVADHDASA-------
**** .*:**:*****:* *****:**: *: :* ::
MUL_3368|M.ulcerans_Agy99 AGSAGAEVPQELLVVNKVDAAGDLALAELRHALPGALFVSAHTGDGIDVL
MMAR_1988|M.marinum_M AGSAGAEAPQELLVVNKVDAAGDLALAELRHALPGALFVSAHTGDGIDVL
MLBr_00997|M.leprae_Br4923 -----VEVPHELLVVNKIDAASALMLAKLRHGLPGAVFISARTGDGIDVL
Mb2744c|M.bovis_AF2122/97 --------PPELLVVNKVDVASDLMLAKLRHGLPGAVFVSARTGDGIDAL
Rv2725c|M.tuberculosis_H37Rv --------PPELLVVNKVDVASDLMLAKLRHGLPGAVFVSARTGDGIDAL
MAV_3618|M.avium_104 --------PPELLVVNKTDAAGDVALAKLRHALPGAVFVSAATGDGIDGL
Mflv_3960|M.gilvum_PYR-GCK --------PPELLVVNKIDAADGLVLAQLRQALPGAVFVSARTGESLDRL
Mvan_2436|M.vanbaalenii_PYR-1 --------PPELLVVNKTDAADGVTLAQLRRALPGAVFVSAHTGDGLDRL
MSMEG_2736|M.smegmatis_MC2_155 --------PPELLVVNKIDAATGVGLAQLRRALPDAVFVSARTGDGLDKL
TH_1520|M.thermoresistible__bu --------APELLVVNKIDAASDLTLTQLRRALPDAVFVSAHTGEGLDRL
MAB_3042c|M.abscessus_ATCC_199 --------APELLVINKVDAAGDLALAQLRRALPKALFVSAHTGEGIATL
. ****:** *.* : *::**:.** *:*:** **:.: *
MUL_3368|M.ulcerans_Agy99 WRKLAELAAPTDTAVDVVIPYDRGDLVARMHADGRVQQAEHQPEGTRIRA
MMAR_1988|M.marinum_M WRKLAELAAPTDTAVDVVIPYDRGDLVARMHADGRVQQAEHQPEGTRIRA
MLBr_00997|M.leprae_Br4923 RRRIAELVVATDTAVDVVIPYDRGDLVARLHANGRVQQAEHNSNGTRIKA
Mb2744c|M.bovis_AF2122/97 RRRMAELVVPADTAVDVVIPYDRGDLVARVHADGRIQQAEHKPEGTRIKA
Rv2725c|M.tuberculosis_H37Rv RRRMAELVVPADTAVDVVIPYDRGDLVARVHADGRIQQAEHKPEGTRIKA
MAV_3618|M.avium_104 RRRIAELAVPAEAAVDVVIPYHRGDLVARLHADGRVQQEEHNAEGTRIKA
Mflv_3960|M.gilvum_PYR-GCK QARLAEMIAPRDTVVDVTIPYSRGDLVNRVHAEGRVDAVEHIEAGTRLKA
Mvan_2436|M.vanbaalenii_PYR-1 RARMAELVVPTDTVVDVTIPYSRGDLVARLHADGRIDATEHTERGTRIKA
MSMEG_2736|M.smegmatis_MC2_155 RSRMGELVESTDATVDVTIPYDRGDLVARVHTDGHVDATEHTDAGTRIKA
TH_1520|M.thermoresistible__bu RQRMSELVAPTETAVDVTIPYDRGDLVARVHSDGRVDATEHRADGTRIKA
MAB_3042c|M.abscessus_ATCC_199 REAIAEAVPRGDVPVDVVIPYERGDLVARIHTEGQVQSTEHLADGTRVVG
:.* :. ***.*** ***** *:*::*::: ** ***: .
MUL_3368|M.ulcerans_Agy99 RVPGALAATLREFAASSAGPGE
MMAR_1988|M.marinum_M RVPGALAATLREFAASPAGPGE
MLBr_00997|M.leprae_Br4923 RVPVALAACLQEFSADR-----
Mb2744c|M.bovis_AF2122/97 RVPEALAATLREFAPRA-----
Rv2725c|M.tuberculosis_H37Rv RVPEALAATLREFAPRA-----
MAV_3618|M.avium_104 RVPLALAGRLREFAAP------
Mflv_3960|M.gilvum_PYR-GCK RVPIALAAALSEFASF------
Mvan_2436|M.vanbaalenii_PYR-1 RVPVSLAAGLADYATY------
MSMEG_2736|M.smegmatis_MC2_155 RVPAPLAATLREYATFA-----
TH_1520|M.thermoresistible__bu RVPVALAASLRDFATV------
MAB_3042c|M.abscessus_ATCC_199 RVPRALAAVLTAL---------
*** .**. *