For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRPDNDPARLRVVAETLAAEAAEFVRSRRAELFGAHPAAADESDAVRAKSTPTDPVTVVDTDAERLLRSR LAQLRPGDPIVGEEGGGPADSAPAPDDAVTWVLDPIDGTVNFVYGIPAYAVSIGAQVGGVSVAGAVADVV GRSVYSAALGQGAHVTDEQGTRELRCTAVEDLSMALLGTGFGYSAGRRATQAALLARMLPMVRDVRRIGS AALDLCMVAAGRLDAYYEHGLQLWDRAAGALIAAEAGARVVLPASDVVGAGLVVAAAPGVADELLGVLKR LNALEPLD*
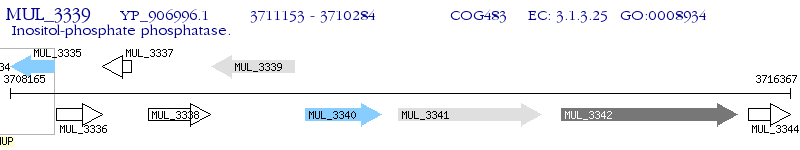
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3339 | suhB | - | 100% (289) | extragenic suppressor protein SuhB |
| M. ulcerans Agy99 | MUL_2430 | - | 2e-18 | 30.97% (226) | monophosphatase |
| M. ulcerans Agy99 | MUL_1577 | impA | 7e-17 | 31.25% (272) | inositol-monophosphatase ImpA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2720c | suhB | 1e-120 | 76.82% (289) | extragenic suppressor protein SuhB |
| M. gilvum PYR-GCK | Mflv_3937 | - | 2e-85 | 61.21% (281) | inositol-phosphate phosphatase |
| M. tuberculosis H37Rv | Rv2701c | suhB | 1e-120 | 76.82% (289) | extragenic suppressor protein SuhB |
| M. leprae Br4923 | MLBr_01024 | suhB | 1e-115 | 75.17% (290) | putative inositol monophosphatase |
| M. abscessus ATCC 19977 | MAB_3007c | - | 6e-79 | 58.25% (285) | inositol-1-monophosphatase SuhB |
| M. marinum M | MMAR_2013 | suhB | 1e-159 | 99.31% (289) | extragenic suppressor protein SuhB |
| M. avium 104 | MAV_3593 | - | 1e-119 | 77.58% (281) | inositol-1-monophosphatase |
| M. smegmatis MC2 155 | MSMEG_2762 | - | 7e-88 | 62.77% (282) | inositol-1-monophosphatase |
| M. thermoresistible (build 8) | TH_0825 | suhB | 2e-79 | 62.25% (249) | POSSIBLE EXTRAGENIC SUPPRESSOR PROTEIN SUHB |
| M. vanbaalenii PYR-1 | Mvan_2463 | - | 2e-90 | 64.41% (281) | inositol-phosphate phosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3339|M.ulcerans_Agy99 -------------------MTRPDNDPARLRVVAETLAAEAAEFVRSRRA
MMAR_2013|M.marinum_M -------------------MTRPDNDPARLRVVAETLAAEAAEFVRSRRA
Mb2720c|M.bovis_AF2122/97 -------------------MTRPDNEPARLRSVAENLAAEAAAFVRGRRA
Rv2701c|M.tuberculosis_H37Rv -------------------MTRPDNEPARLRSVAENLAAEAAAFVRGRRA
MLBr_01024|M.leprae_Br4923 -------------------MTVPDNDPEQLRCVAESLATEAAAFVRCRRA
MAV_3593|M.avium_104 ---------------------------MRLRSVAETLAAEAAAFVRRRRA
MAB_3007c|M.abscessus_ATCC_199 ----------------------MGIEAGALRSVAVRLASEAAEFVRHRRG
Mflv_3937|M.gilvum_PYR-GCK MGPDGRRCAAMDG----VNTMHAADAPVRLQEVAERLAAEAAQFVRRRRV
Mvan_2463|M.vanbaalenii_PYR-1 MGASRASRAARPGRMCCDERVNIDTDPVRLRAVAEQLASEAAEFVGRRRT
TH_0825|M.thermoresistible__bu -------------------MTSAPVDLTELRAVAERLAAEAADFVRRRRA
MSMEG_2762|M.smegmatis_MC2_155 -------------------MTENSTDPAELRAVAEQLADEAAELVLRRRA
*: ** ** *** :* **
MUL_3339|M.ulcerans_Agy99 ELFG---AHPAAADESD----AVRAKSTPTDPVTVVDTDAERLLRSRLAQ
MMAR_2013|M.marinum_M ELFG---AHPAAADESD----AVRAKSTPTDPVTVVDTDAERLLRSRLAQ
Mb2720c|M.bovis_AF2122/97 EVFG---IS-RAGDGDG----AVRAKSSPTDPVTVVDTDTERLLRDRLAQ
Rv2701c|M.tuberculosis_H37Rv EVFG---IS-RAGDGDG----AVRAKSSPTDPVTVVDTDTERLLRDRLAQ
MLBr_01024|M.leprae_Br4923 EVFGTD-LG---AAGGG----AVRAKSTPTDPVTVVDTETERLLRDRLAQ
MAV_3593|M.avium_104 EVFGTE-PGGASAPDGG----AVRSKSTPTDPVTVVDTETERLLRDRLAQ
MAB_3007c|M.abscessus_ATCC_199 -----------TVDWTA----SVRTKSSETDPVTLVDTDCERLLRRRLAS
Mflv_3937|M.gilvum_PYR-GCK EVFGDG-SAGAAPDTAQ---PSVRAKSTPTDPVTIVDTETERLLRDRLGQ
Mvan_2463|M.vanbaalenii_PYR-1 EVFG----AGAAPGGAQ---SAVRAKSSPTDPVTVVDTETERLLRDRLAE
TH_0825|M.thermoresistible__bu EVFGDTGATGASTGSATESGPAVRSKSTPTDPVTIVDTETERLLRDRLTR
MSMEG_2762|M.smegmatis_MC2_155 EVFG---AAGASNDDAAG-GQAVRTKSTPTDPVTVVDTETERWLRERLAV
:**:**: *****:***: ** ** **
MUL_3339|M.ulcerans_Agy99 LRPGDPIVGEEGGGPADSAPAP----DDAVTWVLDPIDGTVNFVYGIPAY
MMAR_2013|M.marinum_M LRPGDPIVGEEGGGPADSAPAP----DDAVTWVLDPIDGTVNFVYGIPAY
Mb2720c|M.bovis_AF2122/97 LRPGDPILGEEGGGPADVTATP----SDRVTWVLDPIDGTVNFVYGIPAY
Rv2701c|M.tuberculosis_H37Rv LRPGDPILGEEGGGPADVTATP----SDRVTWVLDPIDGTVNFVYGIPAY
MLBr_01024|M.leprae_Br4923 LRPGDSILGEEGGGPADLTATP----ADTVTWVLDPIDGTVNFVYGIPAY
MAV_3593|M.avium_104 LRPGEPILGEEGGGPAEPSAPG----DGTVTWVLDPIDGTVNFVYGIPAY
MAB_3007c|M.abscessus_ATCC_199 LRPDDAILGEEDGESAGGSLSGRSGQPSGVRWVIDPIDGTVNFVYGIPAY
Mflv_3937|M.gilvum_PYR-GCK LRPDDAILGEEEGGSVDIAEGR-------LTWVLDPIDGTVNFLYGIPAY
Mvan_2463|M.vanbaalenii_PYR-1 LRPGDAILGEEEGGSVDVADGR-------LTWVLDPIDGTVNFLYGIPAY
TH_0825|M.thermoresistible__bu LRPGEPILGEEGGQSGAAERGR-------PVWVLDPIDGTVNFVYGIEAY
MSMEG_2762|M.smegmatis_MC2_155 LRPGEHVLGEEEGGKQEGRAG--------LSWVLDPIDGTVNFVYGIPAY
***.: ::*** * **:*********:*** **
MUL_3339|M.ulcerans_Agy99 AVSIGAQVGGVSVAGAVADVVGRSVYSAALGQGAHVTDEQGTR-ELRCTA
MMAR_2013|M.marinum_M AVSIGAQVGGVSVAGAVADVVGRSVYSAALGQGAHVTDQQGTR-ELHCTA
Mb2720c|M.bovis_AF2122/97 AVSIGAQVGGITVAGAVADVAARTVYSAATGLGAHLTDERGRH-VLRCTG
Rv2701c|M.tuberculosis_H37Rv AVSIGAQVGGITVAGAVADVAARTVYSAATGLGAHLTDERGRH-VLRCTG
MLBr_01024|M.leprae_Br4923 AVSVAAQVDGVSVAGAVAEVVAGRVHSAASGLGAHVTDEYGVQ-VLRCSA
MAV_3593|M.avium_104 AVSVGAQVDGESVAGAVADVVGDRVYSAAAGLGAHVSDGAGRQ-PLQCAA
MAB_3007c|M.abscessus_ATCC_199 AVSVAAQVGGVTVAGAVADVVADRVFSAASGAGATVREANGLQRPLACNP
Mflv_3937|M.gilvum_PYR-GCK GVSVGVRRDGVSVAGAVADVAAGAVYSAALGHGARLS-RDGVATELHVTS
Mvan_2463|M.vanbaalenii_PYR-1 AVSVGVRRDGVSVAGAVADVAAGAVYSAALGHGAGVR-RGGVLTPLRCTA
TH_0825|M.thermoresistible__bu AVSVGVQIDGESVAGAVADVAAGAVYSAARGHGAHLH-RDGVTRRLRCSA
MSMEG_2762|M.smegmatis_MC2_155 AVSIAVQLDGRSIAGAVAHVSAGAVYSAALGGGAHVR-QNGVTTPLRCSA
.**:..: .* ::*****.* . *.*** * ** : * *
MUL_3339|M.ulcerans_Agy99 VEDLSMALLGTGFGYSAGRRATQAALLARMLPMVRDVRRIGSAALDLCMV
MMAR_2013|M.marinum_M VEDLSMALLGTGFGYSAGRRATQAALLARMLPMVRDVRRIGSAALDLCMV
Mb2720c|M.bovis_AF2122/97 VDELSMALLGTGFGYSVRCREKQAELLAHVVPLVRDVRRIGSAALDLCMV
Rv2701c|M.tuberculosis_H37Rv VDELSMALLGTGFGYSVRCREKQAELLAHVVPLVRDVRRIGSAALDLCMV
MLBr_01024|M.leprae_Br4923 VDDLSMALLGTGFAYSVVRRAAQAALLAQMLPVVRDVRRIGSAALDLCMV
MAV_3593|M.avium_104 VDDVSMALLGTGFGYARQRRAAQAALLARMLPVVRDVRRIGSAALDLCMV
MAB_3007c|M.abscessus_ATCC_199 VRETRLALVATGFAYSTVRRERQAHLVAQLLPRVRDVRRIGSAALDLCAV
Mflv_3937|M.gilvum_PYR-GCK ATDLSMALVGTGFSYDRAHREQQGRIIARLLPEVRDLRRMGSCALDLCAV
Mvan_2463|M.vanbaalenii_PYR-1 ATDLSMALVGTGFSYEREHRQRQGEIAARLLPDVRDVRRMGSCALDLCAV
TH_0825|M.thermoresistible__bu ATELSMALVGTGFAYSAELRRSQARIVATLLPRVRDIRRIGSCALDLCMV
MSMEG_2762|M.smegmatis_MC2_155 VDDLAMTLVGTGFSYSHEARRRQAQVLAQILPTVRDMRRLGSCALDLCMV
. : ::*:.***.* * *. : * ::* ***:**:**.***** *
MUL_3339|M.ulcerans_Agy99 AAGRLDAYYEHGLQLWDRAAGALIAAEAGARVVLPASDV--VGAG-LVVA
MMAR_2013|M.marinum_M AAGRLDAYYEHGLQLWDRAAGALIAAEAGARVVLPASDV--VGAG-LVVA
Mb2720c|M.bovis_AF2122/97 AAGRLDAYYEHGVQVWDCAAGALIAAEAGARVLLSTPRA--GGAGLVVVA
Rv2701c|M.tuberculosis_H37Rv AAGRLDAYYEHGVQVWDCAAGALIAAEAGARVLLSTPRA--GGAGLVVVA
MLBr_01024|M.leprae_Br4923 AAGQLDAYYEHEVQVWDCAAGALIAAEAGACVQLPKRNGPVGGAG-LVVA
MAV_3593|M.avium_104 AAGRLDAYYEHGLKVWDRAAGALIAAEAGARVVLPAPDG--DGAG-LVLA
MAB_3007c|M.abscessus_ATCC_199 AEGRVDAQYEHGLGPWDWAAGALIAREAGAVVILPGPDTR-SAAGALIAA
Mflv_3937|M.gilvum_PYR-GCK AAGRLDAYFEDDVHLWDWAAGALIASEAGALVRPPRTED---GSG-LMVA
Mvan_2463|M.vanbaalenii_PYR-1 AAGRLDAYYEDDVHLWDWAAAALIATEAGAHVRPPATGD---GAG-LMVA
TH_0825|M.thermoresistible__bu AAGLLDAYYEEGVQVWDWAAAXXXVSGGGGGLLGRGAVD---GR------
MSMEG_2762|M.smegmatis_MC2_155 AAGQLDAYYEDGVHVWDWAAAALIAAEAGATVWLP-AGG---GGG-KVIA
* * :** :*. : ** **. . .*. : .
MUL_3339|M.ulcerans_Agy99 AAPGVADELLGVLKRLNALEPLD-
MMAR_2013|M.marinum_M AAPGVADELLGVLKRLNALEPLD-
Mb2720c|M.bovis_AF2122/97 AAPGIADELLAALQRFNGLEPIPD
Rv2701c|M.tuberculosis_H37Rv AAPGIADELLAALQRFNGLEPIPD
MLBr_01024|M.leprae_Br4923 AAPGIADALLAALQRFNGLAPILD
MAV_3593|M.avium_104 AAPGIADELLAVLERFDGLDPILD
MAB_3007c|M.abscessus_ATCC_199 TAPGIAGEFGQLLADIGATAPIPG
Mflv_3937|M.gilvum_PYR-GCK AAPGISVALDEALARSGAL-----
Mvan_2463|M.vanbaalenii_PYR-1 AAPGISAAFDAALIRCGAVGG---
TH_0825|M.thermoresistible__bu -------------ARRGAGR----
MSMEG_2762|M.smegmatis_MC2_155 AAPGVATALRDALAGAGMDL----
.